Régression polynômiale et pipeline¶
Le notebook compare plusieurs de modèles de régression polynômiale.
[2]:
%matplotlib inline
[3]:
from teachpyx.datasets import load_wines_dataset
data = load_wines_dataset()
X = data.drop(["quality", "color"], axis=1)
y = data["quality"]
[4]:
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y)
On normalise les données. Pour ce cas particulier, c’est d’autant plus important que les polynômes prendront de très grandes valeurs si cela n’est pas fait et les librairies de calculs n’aiment pas les ordres de grandeurs trop différents.
[5]:
from sklearn.preprocessing import Normalizer
norm = Normalizer()
X_train_norm = norm.fit_transform(X_train)
X_test_norm = norm.transform(X_test)
La transformation PolynomialFeatures créée de nouvelles features en multipliant les variables les unes avec les autres. Pour le degré deux et trois features , on obtient les nouvelles features :
.
[6]:
from time import perf_counter
from sklearn.linear_model import LinearRegression
from sklearn.preprocessing import PolynomialFeatures
from sklearn.pipeline import make_pipeline
from sklearn.metrics import r2_score
r2ts = []
r2es = []
degs = []
tts = []
models = []
for d in range(1, 5):
begin = perf_counter()
pipe = make_pipeline(PolynomialFeatures(degree=d), LinearRegression())
pipe.fit(X_train_norm, y_train)
duree = perf_counter() - begin
r2t = r2_score(y_train, pipe.predict(X_train_norm))
r2e = r2_score(y_test, pipe.predict(X_test_norm))
degs.append(d)
r2ts.append(r2t)
r2es.append(r2e)
tts.append(duree)
models.append(pipe)
print(d, r2t, r2e, duree)
1 0.1909065078664849 0.16570749381482386 0.02195639999990817
2 0.31686272332465504 0.2634484656108902 0.16658860000006825
3 0.4117084105383497 -1.446755311176299 1.0382120000001578
4 0.5940872457783092 -3926.677572477097 2.8583189999999377
[7]:
import pandas
df = pandas.DataFrame(dict(temps=tts, r2_train=r2ts, r2_test=r2es, degré=degs))
df.set_index("degré")
[7]:
| temps | r2_train | r2_test | |
|---|---|---|---|
| degré | |||
| 1 | 0.021956 | 0.190907 | 0.165707 |
| 2 | 0.166589 | 0.316863 | 0.263448 |
| 3 | 1.038212 | 0.411708 | -1.446755 |
| 4 | 2.858319 | 0.594087 | -3926.677572 |
Le polynômes de degré 2 paraît le meilleur modèle. Le temps de calcul est multiplié par 10 à chaque fois, ce qui correspond au nombre de features. On voit néanmoins que l’ajout de features croisée fonctionne sur ce jeu de données. Mais au delà de 3, la régression produit des résultats très mauvais sur la base de test alors qu’ils continuent d’augmenter sur la base d’apprentissage. Voyons cela un peu plus en détail.
[7]:
import matplotlib.pyplot as plt
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
n = 15
ax[0].plot(y_train[:n].reset_index(), "o")
ax[1].plot(y_test[:n].reset_index(), "o")
ax[0].set_title("Prédictions sur quelques valeurs\napprentissage")
ax[1].set_title("Prédictions sur quelques valeurs\ntest")
for x in ax:
x.set_ylim([3, 9])
x.get_xaxis().set_visible(False)
for model in models:
d = model.get_params()["polynomialfeatures__degree"]
tr = model.predict(X_train_norm[:n])
te = model.predict(X_test_norm[:n])
ax[0].plot(tr, label="d=%d" % d)
ax[1].plot(te, label="d=%d" % d)
ax[0].legend()
ax[1].legend();
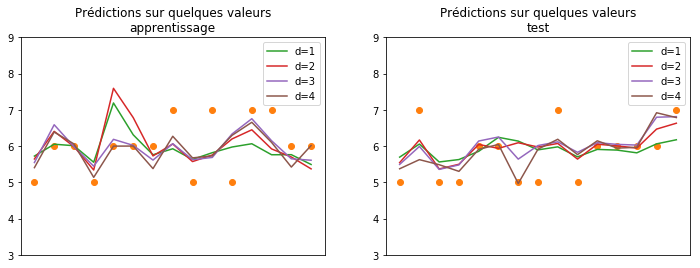
Le modèle de degré 4 a l’air performant sur la base d’apprentissage mais s’égare complètement sur la base de test comme s’il était surpris des valeurs rencontrées sur la base de test. On dit que le modèle fait du sur-apprentissage ou overfitting en anglais. Le polynôme de degré fonctionne mieux que la régression linéaire simple. On peut se demander quelles sont les variables croisées qui ont un impact sur la performance. On utilise le modèle statsmodels.
[8]:
poly = PolynomialFeatures(degree=2)
poly_feat_train = poly.fit_transform(X_train_norm)
poly_feat_test = poly.fit_transform(X_test_norm)
[9]:
from statsmodels.regression.linear_model import OLS
model = OLS(y_train, poly_feat_train)
results = model.fit()
results.summary2()
[9]:
| Model: | OLS | Adj. R-squared: | 0.306 |
| Dependent Variable: | quality | AIC: | 10768.5223 |
| Date: | 2024-01-23 00:08 | BIC: | 11268.3493 |
| No. Observations: | 4872 | Log-Likelihood: | -5307.3 |
| Df Model: | 76 | F-statistic: | 29.30 |
| Df Residuals: | 4795 | Prob (F-statistic): | 0.00 |
| R-squared: | 0.317 | Scale: | 0.52557 |
| Coef. | Std.Err. | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| const | 874.2126 | 1866.6100 | 0.4683 | 0.6396 | -2785.1996 | 4533.6248 |
| x1 | 17.2438 | 25.1175 | 0.6865 | 0.4924 | -31.9980 | 66.4856 |
| x2 | -735.9147 | 164.2593 | -4.4802 | 0.0000 | -1057.9383 | -413.8911 |
| x3 | -375.2205 | 200.9788 | -1.8670 | 0.0620 | -769.2311 | 18.7900 |
| x4 | 2.1457 | 13.7859 | 0.1556 | 0.8763 | -24.8809 | 29.1723 |
| x5 | -1219.9140 | 760.0849 | -1.6050 | 0.1086 | -2710.0291 | 270.2011 |
| x6 | 33.0684 | 8.6300 | 3.8318 | 0.0001 | 16.1496 | 49.9873 |
| x7 | 45.6122 | 23.6785 | 1.9263 | 0.0541 | -0.8085 | 92.0328 |
| x8 | -1621.7821 | 721.4602 | -2.2479 | 0.0246 | -3036.1752 | -207.3890 |
| x9 | -123.6719 | 196.5043 | -0.6294 | 0.5291 | -508.9104 | 261.5667 |
| x10 | -213.6188 | 172.6441 | -1.2373 | 0.2160 | -552.0806 | 124.8429 |
| x11 | 274.6811 | 25.3731 | 10.8257 | 0.0000 | 224.9381 | 324.4241 |
| x12 | -888.0924 | 1860.0506 | -0.4775 | 0.6331 | -4534.6449 | 2758.4602 |
| x13 | 213.0448 | 149.3410 | 1.4266 | 0.1538 | -79.7320 | 505.8216 |
| x14 | -169.2454 | 191.2389 | -0.8850 | 0.3762 | -544.1614 | 205.6706 |
| x15 | -2.3959 | 21.0911 | -0.1136 | 0.9096 | -43.7441 | 38.9523 |
| x16 | 151.4367 | 661.2643 | 0.2290 | 0.8189 | -1144.9447 | 1447.8180 |
| x17 | -13.3943 | 9.8122 | -1.3651 | 0.1723 | -32.6306 | 5.8421 |
| x18 | -12.3144 | 22.1599 | -0.5557 | 0.5784 | -55.7580 | 31.1291 |
| x19 | -228.1023 | 1055.2972 | -0.2161 | 0.8289 | -2296.9691 | 1840.7644 |
| x20 | 263.5729 | 260.8085 | 1.0106 | 0.3123 | -247.7314 | 774.8773 |
| x21 | 210.1261 | 147.0152 | 1.4293 | 0.1530 | -78.0912 | 498.3434 |
| x22 | -102.4573 | 26.1357 | -3.9202 | 0.0001 | -153.6952 | -51.2193 |
| x23 | -1256.2263 | 1979.4050 | -0.6346 | 0.5257 | -5136.7684 | 2624.3158 |
| x24 | 2503.6940 | 1629.0043 | 1.5369 | 0.1244 | -689.9019 | 5697.2899 |
| x25 | -304.5840 | 139.4970 | -2.1834 | 0.0291 | -578.0621 | -31.1060 |
| x26 | 6503.3193 | 4276.6479 | 1.5207 | 0.1284 | -1880.8729 | 14887.5116 |
| x27 | 176.1465 | 65.4348 | 2.6919 | 0.0071 | 47.8643 | 304.4288 |
| x28 | 541.9165 | 145.6814 | 3.7199 | 0.0002 | 256.3141 | 827.5190 |
| x29 | -5408.0462 | 5170.0682 | -1.0460 | 0.2956 | -15543.7521 | 4727.6598 |
| x30 | 1591.6613 | 1576.5785 | 1.0096 | 0.3128 | -1499.1559 | 4682.4786 |
| x31 | -3066.2318 | 1347.2069 | -2.2760 | 0.0229 | -5707.3755 | -425.0882 |
| x32 | 611.6934 | 183.2814 | 3.3375 | 0.0009 | 252.3778 | 971.0089 |
| x33 | 861.9664 | 2070.4337 | 0.4163 | 0.6772 | -3197.0336 | 4920.9664 |
| x34 | -307.3877 | 171.8498 | -1.7887 | 0.0737 | -644.2921 | 29.5168 |
| x35 | -8483.4913 | 6547.3948 | -1.2957 | 0.1951 | -21319.3893 | 4352.4067 |
| x36 | 150.5489 | 83.1030 | 1.8116 | 0.0701 | -12.3711 | 313.4689 |
| x37 | 300.7497 | 178.4947 | 1.6849 | 0.0921 | -49.1819 | 650.6813 |
| x38 | 14067.7740 | 7800.2113 | 1.8035 | 0.0714 | -1224.2191 | 29359.7672 |
| x39 | -5133.5558 | 2077.6861 | -2.4708 | 0.0135 | -9206.7738 | -1060.3378 |
| x40 | -2372.2746 | 1576.8448 | -1.5044 | 0.1325 | -5463.6139 | 719.0647 |
| x41 | 708.8006 | 236.3385 | 2.9991 | 0.0027 | 245.4687 | 1172.1325 |
| x42 | -910.1293 | 1867.0943 | -0.4875 | 0.6260 | -4570.4908 | 2750.2323 |
| x43 | 1971.4865 | 757.4887 | 2.6027 | 0.0093 | 486.4611 | 3456.5118 |
| x44 | -7.6328 | 5.0273 | -1.5183 | 0.1290 | -17.4886 | 2.2230 |
| x45 | 2.8665 | 12.6000 | 0.2275 | 0.8200 | -21.8354 | 27.5684 |
| x46 | 1429.2194 | 705.7754 | 2.0250 | 0.0429 | 45.5757 | 2812.8631 |
| x47 | -287.7160 | 203.0709 | -1.4168 | 0.1566 | -685.8281 | 110.3961 |
| x48 | -189.7045 | 168.1916 | -1.1279 | 0.2594 | -519.4371 | 140.0282 |
| x49 | -18.0129 | 19.5540 | -0.9212 | 0.3570 | -56.3477 | 20.3219 |
| x50 | 10142.2002 | 7074.9790 | 1.4335 | 0.1518 | -3728.0049 | 24012.4054 |
| x51 | -201.9536 | 331.1123 | -0.6099 | 0.5419 | -851.0857 | 447.1785 |
| x52 | 1197.2260 | 682.4747 | 1.7542 | 0.0795 | -140.7375 | 2535.1896 |
| x53 | 20265.9335 | 23064.5613 | 0.8787 | 0.3796 | -24951.1898 | 65483.0568 |
| x54 | -12226.8108 | 6517.5063 | -1.8760 | 0.0607 | -25004.1137 | 550.4920 |
| x55 | -3613.4768 | 4978.7999 | -0.7258 | 0.4680 | -13374.2092 | 6147.2555 |
| x56 | 2196.0436 | 843.4185 | 2.6037 | 0.0092 | 542.5563 | 3849.5310 |
| x57 | -909.6884 | 1867.1743 | -0.4872 | 0.6261 | -4570.2067 | 2750.8299 |
| x58 | -24.9437 | 7.6926 | -3.2426 | 0.0012 | -40.0247 | -9.8627 |
| x59 | 549.2329 | 298.2374 | 1.8416 | 0.0656 | -35.4492 | 1133.9150 |
| x60 | -13.8185 | 79.9507 | -0.1728 | 0.8628 | -170.5585 | 142.9216 |
| x61 | 59.9612 | 69.6414 | 0.8610 | 0.3893 | -76.5679 | 196.4903 |
| x62 | -60.9958 | 9.6344 | -6.3310 | 0.0000 | -79.8838 | -42.1079 |
| x63 | -915.2771 | 1867.4927 | -0.4901 | 0.6241 | -4576.4197 | 2745.8655 |
| x64 | 856.3795 | 643.6237 | 1.3306 | 0.1834 | -405.4183 | 2118.1773 |
| x65 | 172.8750 | 176.1471 | 0.9814 | 0.3264 | -172.4541 | 518.2041 |
| x66 | 233.8040 | 154.5694 | 1.5126 | 0.1304 | -69.2230 | 536.8310 |
| x67 | -208.7613 | 22.8714 | -9.1276 | 0.0000 | -253.5998 | -163.9229 |
| x68 | 2377.5655 | 18554.7387 | 0.1281 | 0.8980 | -33998.2360 | 38753.3671 |
| x69 | 11008.3563 | 10291.6307 | 1.0696 | 0.2848 | -9167.9622 | 31184.6748 |
| x70 | -6367.0944 | 6065.2219 | -1.0498 | 0.2939 | -18257.7123 | 5523.5234 |
| x71 | -2160.1700 | 944.6336 | -2.2868 | 0.0223 | -4012.0853 | -308.2546 |
| x72 | -4031.2392 | 1137.0986 | -3.5452 | 0.0004 | -6260.4741 | -1802.0043 |
| x73 | 1604.8538 | 1639.6859 | 0.9788 | 0.3277 | -1609.6830 | 4819.3905 |
| x74 | 766.9861 | 249.3365 | 3.0761 | 0.0021 | 278.1721 | 1255.8001 |
| x75 | -2563.1016 | 2045.6050 | -1.2530 | 0.2103 | -6573.4261 | 1447.2228 |
| x76 | 596.8511 | 210.7614 | 2.8319 | 0.0046 | 183.6620 | 1010.0402 |
| x77 | -1033.7653 | 1865.5885 | -0.5541 | 0.5795 | -4691.1748 | 2623.6442 |
| Omnibus: | 74.655 | Durbin-Watson: | 2.012 |
| Prob(Omnibus): | 0.000 | Jarque-Bera (JB): | 119.025 |
| Skew: | 0.141 | Prob(JB): | 0.000 |
| Kurtosis: | 3.712 | Condition No.: | 4390541685726112 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[2] The smallest eigenvalue is 7.09e-28. This might indicate that there are strong multicollinearity problems or that the design matrix is singular.
Ce n’est pas très lisible. Il faut ajouter le nom de chaque variable et recommencer.
[12]:
names = poly.get_feature_names_out(input_features=data.columns[:-2])
names = [n.replace(" ", " * ") for n in names]
pft = pandas.DataFrame(poly_feat_train, columns=names)
pft.head()
[12]:
| 1 | fixed_acidity | volatile_acidity | citric_acid | residual_sugar | chlorides | free_sulfur_dioxide | total_sulfur_dioxide | density | pH | ... | density^2 | density * pH | density * sulphates | density * alcohol | pH^2 | pH * sulphates | pH * alcohol | sulphates^2 | sulphates * alcohol | alcohol^2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.0 | 0.061316 | 0.003511 | 0.003461 | 0.019779 | 0.000455 | 0.306582 | 0.939526 | 0.009773 | 0.030263 | ... | 0.000096 | 0.000296 | 0.000044 | 0.001315 | 0.000916 | 0.000138 | 0.004070 | 0.000021 | 0.000612 | 0.018090 |
| 1 | 1.0 | 0.035236 | 0.001373 | 0.001922 | 0.065438 | 0.000206 | 0.205924 | 0.974706 | 0.004572 | 0.014552 | ... | 0.000021 | 0.000067 | 0.000013 | 0.000192 | 0.000212 | 0.000042 | 0.000613 | 0.000008 | 0.000121 | 0.001772 |
| 2 | 1.0 | 0.042579 | 0.001319 | 0.001919 | 0.101349 | 0.000336 | 0.293852 | 0.947522 | 0.005996 | 0.020210 | ... | 0.000036 | 0.000121 | 0.000014 | 0.000345 | 0.000408 | 0.000046 | 0.001163 | 0.000005 | 0.000131 | 0.003314 |
| 3 | 1.0 | 0.053638 | 0.000920 | 0.001456 | 0.037547 | 0.000421 | 0.206890 | 0.973147 | 0.007627 | 0.025210 | ... | 0.000058 | 0.000192 | 0.000024 | 0.000549 | 0.000636 | 0.000079 | 0.001816 | 0.000010 | 0.000226 | 0.005188 |
| 4 | 1.0 | 0.071498 | 0.002413 | 0.002949 | 0.010725 | 0.000447 | 0.366428 | 0.920540 | 0.008848 | 0.026812 | ... | 0.000078 | 0.000237 | 0.000036 | 0.000981 | 0.000719 | 0.000108 | 0.002971 | 0.000016 | 0.000446 | 0.012282 |
5 rows × 78 columns
[13]:
results.summary2(xname=pft.columns)
[13]:
| Model: | OLS | Adj. R-squared: | 0.306 |
| Dependent Variable: | quality | AIC: | 10768.5223 |
| Date: | 2024-01-23 00:09 | BIC: | 11268.3493 |
| No. Observations: | 4872 | Log-Likelihood: | -5307.3 |
| Df Model: | 76 | F-statistic: | 29.30 |
| Df Residuals: | 4795 | Prob (F-statistic): | 0.00 |
| R-squared: | 0.317 | Scale: | 0.52557 |
| Coef. | Std.Err. | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| 1 | 874.2126 | 1866.6100 | 0.4683 | 0.6396 | -2785.1996 | 4533.6248 |
| fixed_acidity | 17.2438 | 25.1175 | 0.6865 | 0.4924 | -31.9980 | 66.4856 |
| volatile_acidity | -735.9147 | 164.2593 | -4.4802 | 0.0000 | -1057.9383 | -413.8911 |
| citric_acid | -375.2205 | 200.9788 | -1.8670 | 0.0620 | -769.2311 | 18.7900 |
| residual_sugar | 2.1457 | 13.7859 | 0.1556 | 0.8763 | -24.8809 | 29.1723 |
| chlorides | -1219.9140 | 760.0849 | -1.6050 | 0.1086 | -2710.0291 | 270.2011 |
| free_sulfur_dioxide | 33.0684 | 8.6300 | 3.8318 | 0.0001 | 16.1496 | 49.9873 |
| total_sulfur_dioxide | 45.6122 | 23.6785 | 1.9263 | 0.0541 | -0.8085 | 92.0328 |
| density | -1621.7821 | 721.4602 | -2.2479 | 0.0246 | -3036.1752 | -207.3890 |
| pH | -123.6719 | 196.5043 | -0.6294 | 0.5291 | -508.9104 | 261.5667 |
| sulphates | -213.6188 | 172.6441 | -1.2373 | 0.2160 | -552.0806 | 124.8429 |
| alcohol | 274.6811 | 25.3731 | 10.8257 | 0.0000 | 224.9381 | 324.4241 |
| fixed_acidity^2 | -888.0924 | 1860.0506 | -0.4775 | 0.6331 | -4534.6449 | 2758.4602 |
| fixed_acidity * volatile_acidity | 213.0448 | 149.3410 | 1.4266 | 0.1538 | -79.7320 | 505.8216 |
| fixed_acidity * citric_acid | -169.2454 | 191.2389 | -0.8850 | 0.3762 | -544.1614 | 205.6706 |
| fixed_acidity * residual_sugar | -2.3959 | 21.0911 | -0.1136 | 0.9096 | -43.7441 | 38.9523 |
| fixed_acidity * chlorides | 151.4367 | 661.2643 | 0.2290 | 0.8189 | -1144.9447 | 1447.8180 |
| fixed_acidity * free_sulfur_dioxide | -13.3943 | 9.8122 | -1.3651 | 0.1723 | -32.6306 | 5.8421 |
| fixed_acidity * total_sulfur_dioxide | -12.3144 | 22.1599 | -0.5557 | 0.5784 | -55.7580 | 31.1291 |
| fixed_acidity * density | -228.1023 | 1055.2972 | -0.2161 | 0.8289 | -2296.9691 | 1840.7644 |
| fixed_acidity * pH | 263.5729 | 260.8085 | 1.0106 | 0.3123 | -247.7314 | 774.8773 |
| fixed_acidity * sulphates | 210.1261 | 147.0152 | 1.4293 | 0.1530 | -78.0912 | 498.3434 |
| fixed_acidity * alcohol | -102.4573 | 26.1357 | -3.9202 | 0.0001 | -153.6952 | -51.2193 |
| volatile_acidity^2 | -1256.2263 | 1979.4050 | -0.6346 | 0.5257 | -5136.7684 | 2624.3158 |
| volatile_acidity * citric_acid | 2503.6940 | 1629.0043 | 1.5369 | 0.1244 | -689.9019 | 5697.2899 |
| volatile_acidity * residual_sugar | -304.5840 | 139.4970 | -2.1834 | 0.0291 | -578.0621 | -31.1060 |
| volatile_acidity * chlorides | 6503.3193 | 4276.6479 | 1.5207 | 0.1284 | -1880.8729 | 14887.5116 |
| volatile_acidity * free_sulfur_dioxide | 176.1465 | 65.4348 | 2.6919 | 0.0071 | 47.8643 | 304.4288 |
| volatile_acidity * total_sulfur_dioxide | 541.9165 | 145.6814 | 3.7199 | 0.0002 | 256.3141 | 827.5190 |
| volatile_acidity * density | -5408.0462 | 5170.0682 | -1.0460 | 0.2956 | -15543.7521 | 4727.6598 |
| volatile_acidity * pH | 1591.6613 | 1576.5785 | 1.0096 | 0.3128 | -1499.1559 | 4682.4786 |
| volatile_acidity * sulphates | -3066.2318 | 1347.2069 | -2.2760 | 0.0229 | -5707.3755 | -425.0882 |
| volatile_acidity * alcohol | 611.6934 | 183.2814 | 3.3375 | 0.0009 | 252.3778 | 971.0089 |
| citric_acid^2 | 861.9664 | 2070.4337 | 0.4163 | 0.6772 | -3197.0336 | 4920.9664 |
| citric_acid * residual_sugar | -307.3877 | 171.8498 | -1.7887 | 0.0737 | -644.2921 | 29.5168 |
| citric_acid * chlorides | -8483.4913 | 6547.3948 | -1.2957 | 0.1951 | -21319.3893 | 4352.4067 |
| citric_acid * free_sulfur_dioxide | 150.5489 | 83.1030 | 1.8116 | 0.0701 | -12.3711 | 313.4689 |
| citric_acid * total_sulfur_dioxide | 300.7497 | 178.4947 | 1.6849 | 0.0921 | -49.1819 | 650.6813 |
| citric_acid * density | 14067.7740 | 7800.2113 | 1.8035 | 0.0714 | -1224.2191 | 29359.7672 |
| citric_acid * pH | -5133.5558 | 2077.6861 | -2.4708 | 0.0135 | -9206.7738 | -1060.3378 |
| citric_acid * sulphates | -2372.2746 | 1576.8448 | -1.5044 | 0.1325 | -5463.6139 | 719.0647 |
| citric_acid * alcohol | 708.8006 | 236.3385 | 2.9991 | 0.0027 | 245.4687 | 1172.1325 |
| residual_sugar^2 | -910.1293 | 1867.0943 | -0.4875 | 0.6260 | -4570.4908 | 2750.2323 |
| residual_sugar * chlorides | 1971.4865 | 757.4887 | 2.6027 | 0.0093 | 486.4611 | 3456.5118 |
| residual_sugar * free_sulfur_dioxide | -7.6328 | 5.0273 | -1.5183 | 0.1290 | -17.4886 | 2.2230 |
| residual_sugar * total_sulfur_dioxide | 2.8665 | 12.6000 | 0.2275 | 0.8200 | -21.8354 | 27.5684 |
| residual_sugar * density | 1429.2194 | 705.7754 | 2.0250 | 0.0429 | 45.5757 | 2812.8631 |
| residual_sugar * pH | -287.7160 | 203.0709 | -1.4168 | 0.1566 | -685.8281 | 110.3961 |
| residual_sugar * sulphates | -189.7045 | 168.1916 | -1.1279 | 0.2594 | -519.4371 | 140.0282 |
| residual_sugar * alcohol | -18.0129 | 19.5540 | -0.9212 | 0.3570 | -56.3477 | 20.3219 |
| chlorides^2 | 10142.2002 | 7074.9790 | 1.4335 | 0.1518 | -3728.0049 | 24012.4054 |
| chlorides * free_sulfur_dioxide | -201.9536 | 331.1123 | -0.6099 | 0.5419 | -851.0857 | 447.1785 |
| chlorides * total_sulfur_dioxide | 1197.2260 | 682.4747 | 1.7542 | 0.0795 | -140.7375 | 2535.1896 |
| chlorides * density | 20265.9335 | 23064.5613 | 0.8787 | 0.3796 | -24951.1898 | 65483.0568 |
| chlorides * pH | -12226.8108 | 6517.5063 | -1.8760 | 0.0607 | -25004.1137 | 550.4920 |
| chlorides * sulphates | -3613.4768 | 4978.7999 | -0.7258 | 0.4680 | -13374.2092 | 6147.2555 |
| chlorides * alcohol | 2196.0436 | 843.4185 | 2.6037 | 0.0092 | 542.5563 | 3849.5310 |
| free_sulfur_dioxide^2 | -909.6884 | 1867.1743 | -0.4872 | 0.6261 | -4570.2067 | 2750.8299 |
| free_sulfur_dioxide * total_sulfur_dioxide | -24.9437 | 7.6926 | -3.2426 | 0.0012 | -40.0247 | -9.8627 |
| free_sulfur_dioxide * density | 549.2329 | 298.2374 | 1.8416 | 0.0656 | -35.4492 | 1133.9150 |
| free_sulfur_dioxide * pH | -13.8185 | 79.9507 | -0.1728 | 0.8628 | -170.5585 | 142.9216 |
| free_sulfur_dioxide * sulphates | 59.9612 | 69.6414 | 0.8610 | 0.3893 | -76.5679 | 196.4903 |
| free_sulfur_dioxide * alcohol | -60.9958 | 9.6344 | -6.3310 | 0.0000 | -79.8838 | -42.1079 |
| total_sulfur_dioxide^2 | -915.2771 | 1867.4927 | -0.4901 | 0.6241 | -4576.4197 | 2745.8655 |
| total_sulfur_dioxide * density | 856.3795 | 643.6237 | 1.3306 | 0.1834 | -405.4183 | 2118.1773 |
| total_sulfur_dioxide * pH | 172.8750 | 176.1471 | 0.9814 | 0.3264 | -172.4541 | 518.2041 |
| total_sulfur_dioxide * sulphates | 233.8040 | 154.5694 | 1.5126 | 0.1304 | -69.2230 | 536.8310 |
| total_sulfur_dioxide * alcohol | -208.7613 | 22.8714 | -9.1276 | 0.0000 | -253.5998 | -163.9229 |
| density^2 | 2377.5655 | 18554.7387 | 0.1281 | 0.8980 | -33998.2360 | 38753.3671 |
| density * pH | 11008.3563 | 10291.6307 | 1.0696 | 0.2848 | -9167.9622 | 31184.6748 |
| density * sulphates | -6367.0944 | 6065.2219 | -1.0498 | 0.2939 | -18257.7123 | 5523.5234 |
| density * alcohol | -2160.1700 | 944.6336 | -2.2868 | 0.0223 | -4012.0853 | -308.2546 |
| pH^2 | -4031.2392 | 1137.0986 | -3.5452 | 0.0004 | -6260.4741 | -1802.0043 |
| pH * sulphates | 1604.8538 | 1639.6859 | 0.9788 | 0.3277 | -1609.6830 | 4819.3905 |
| pH * alcohol | 766.9861 | 249.3365 | 3.0761 | 0.0021 | 278.1721 | 1255.8001 |
| sulphates^2 | -2563.1016 | 2045.6050 | -1.2530 | 0.2103 | -6573.4261 | 1447.2228 |
| sulphates * alcohol | 596.8511 | 210.7614 | 2.8319 | 0.0046 | 183.6620 | 1010.0402 |
| alcohol^2 | -1033.7653 | 1865.5885 | -0.5541 | 0.5795 | -4691.1748 | 2623.6442 |
| Omnibus: | 74.655 | Durbin-Watson: | 2.012 |
| Prob(Omnibus): | 0.000 | Jarque-Bera (JB): | 119.025 |
| Skew: | 0.141 | Prob(JB): | 0.000 |
| Kurtosis: | 3.712 | Condition No.: | 4390541685726112 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[2] The smallest eigenvalue is 7.09e-28. This might indicate that there are strong multicollinearity problems or that the design matrix is singular.
On ne garde que celles dont la p-value est inférieur à 0.05.
[14]:
pval = results.pvalues.copy()
pval[pval <= 0.05]
[14]:
x2 7.630773e-06
x6 1.288486e-04
x8 2.462679e-02
x11 5.327338e-27
x22 8.970958e-05
x25 2.905133e-02
x27 7.128466e-03
x28 2.016020e-04
x31 2.289041e-02
x32 8.519337e-04
x39 1.351547e-02
x41 2.721775e-03
x43 9.278805e-03
x46 4.291916e-02
x56 9.249665e-03
x58 1.192719e-03
x62 2.657965e-10
x67 1.010018e-19
x71 2.225201e-02
x72 3.960632e-04
x74 2.109039e-03
x76 4.646809e-03
dtype: float64
[15]:
pval.index = pft.columns
pval[pval <= 0.05]
[15]:
volatile_acidity 7.630773e-06
free_sulfur_dioxide 1.288486e-04
density 2.462679e-02
alcohol 5.327338e-27
fixed_acidity * alcohol 8.970958e-05
volatile_acidity * residual_sugar 2.905133e-02
volatile_acidity * free_sulfur_dioxide 7.128466e-03
volatile_acidity * total_sulfur_dioxide 2.016020e-04
volatile_acidity * sulphates 2.289041e-02
volatile_acidity * alcohol 8.519337e-04
citric_acid * pH 1.351547e-02
citric_acid * alcohol 2.721775e-03
residual_sugar * chlorides 9.278805e-03
residual_sugar * density 4.291916e-02
chlorides * alcohol 9.249665e-03
free_sulfur_dioxide * total_sulfur_dioxide 1.192719e-03
free_sulfur_dioxide * alcohol 2.657965e-10
total_sulfur_dioxide * alcohol 1.010018e-19
density * alcohol 2.225201e-02
pH^2 3.960632e-04
pH * alcohol 2.109039e-03
sulphates * alcohol 4.646809e-03
dtype: float64
Le modèle fonctionne mieux mais il est plus compliqué de savoir si la contribution de l’alcool est corrélée positivement avec la qualité car l’alcool apparaît dans plus d’une variable.