Prédiction de la note des vins¶
Le notebook compare plusieurs de modèles de régression.
[13]:
%matplotlib inline
[14]:
import warnings
warnings.simplefilter("ignore")
[15]:
from teachpyx.datasets import load_wines_dataset
df = load_wines_dataset()
X = df.drop(["quality", "color"], axis=1)
y = yn = df["quality"]
[16]:
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y)
On normalise les données.
[17]:
from sklearn.preprocessing import normalize
X_train_norm = normalize(X_train)
X_test_norm = normalize(X_test)
X_norm = normalize(X)
Cette façon de faire est complètement erronnée car il est peu probable que la même normalisation soit apppliquée sur les trois bases. La normalisation doit être estimée sur la base d’apprentissage et appliquée sur la base de test. Reprenons.
[18]:
from sklearn.preprocessing import Normalizer
norm = Normalizer()
X_train_norm = norm.fit_transform(X_train)
X_test_norm = norm.transform(X_test)
X_norm = norm.transform(X)
On récupère beaucoup de modèles implémentés dans scikit-learn pour la régression.
[19]:
from sklearn.linear_model import *
from sklearn.ensemble import *
from sklearn.neighbors import *
from sklearn.neural_network import *
models = [
model
for name, model in globals().items()
if (
hasattr(model, "fit")
and not hasattr(model, "predict_proba")
and hasattr(model, "__name__")
and not model.__name__.endswith("CV")
and "Logistic" not in model.__name__
and "Regr" in model.__name__
)
]
import pprint
pprint.pprint(models)
[<class 'sklearn.linear_model._bayes.ARDRegression'>,
<class 'sklearn.linear_model._huber.HuberRegressor'>,
<class 'sklearn.linear_model._base.LinearRegression'>,
<class 'sklearn.linear_model._passive_aggressive.PassiveAggressiveRegressor'>,
<class 'sklearn.linear_model._quantile.QuantileRegressor'>,
<class 'sklearn.linear_model._stochastic_gradient.SGDRegressor'>,
<class 'sklearn.linear_model._theil_sen.TheilSenRegressor'>,
<class 'sklearn.linear_model._ransac.RANSACRegressor'>,
<class 'sklearn.linear_model._glm.glm.PoissonRegressor'>,
<class 'sklearn.linear_model._glm.glm.GammaRegressor'>,
<class 'sklearn.linear_model._glm.glm.TweedieRegressor'>,
<class 'sklearn.ensemble._forest.RandomForestRegressor'>,
<class 'sklearn.ensemble._forest.ExtraTreesRegressor'>,
<class 'sklearn.ensemble._bagging.BaggingRegressor'>,
<class 'sklearn.ensemble._gb.GradientBoostingRegressor'>,
<class 'sklearn.ensemble._weight_boosting.AdaBoostRegressor'>,
<class 'sklearn.ensemble._voting.VotingRegressor'>,
<class 'sklearn.ensemble._stacking.StackingRegressor'>,
<class 'sklearn.ensemble._hist_gradient_boosting.gradient_boosting.HistGradientBoostingRegressor'>,
<class 'sklearn.neighbors._regression.KNeighborsRegressor'>,
<class 'sklearn.neighbors._regression.RadiusNeighborsRegressor'>,
<class 'sklearn.neural_network._multilayer_perceptron.MLPRegressor'>]
[20]:
from sklearn.metrics import r2_score
def score_model(xtr, xte, ytr, yte, model):
try:
model.fit(xtr, ytr)
except Exception as e:
raise Exception("Issue with model '{0}'".format(model.__name__)) from e
return r2_score(yte, model.predict(xte))
[21]:
from time import perf_counter
r2s = []
names = []
durations = []
regressors = {}
for i, model in enumerate(models):
if model.__name__ in {
"ARDRegression",
"VotingRegressor",
"StackingRegressor",
"QuantileRegressor",
}:
continue
try:
reg = model()
except Exception as e:
print("Skip", model)
continue
begin = perf_counter()
r2 = score_model(X_train_norm, X_test_norm, y_train, y_test, reg)
duree = perf_counter() - begin
r2s.append(r2)
names.append(model.__name__)
durations.append(duree)
regressors[model.__name__] = reg
print(i, model.__name__, r2, duree)
1 HuberRegressor 0.15264869419166682 1.0747132999999849
2 LinearRegression 0.17989904732301487 0.029094200000145065
3 PassiveAggressiveRegressor -0.06283962040801283 0.022322499999972933
5 SGDRegressor 0.008293681864454783 0.019755700000132492
6 TheilSenRegressor -0.29442406131949195 1.6061727999999675
7 RANSACRegressor 0.09980597599411645 0.2597002999996221
8 PoissonRegressor 0.0013198388054543875 0.09024569999974119
9 GammaRegressor -0.00011329963875650328 0.02906749999965541
10 TweedieRegressor -0.00012663753861930083 0.06156760000021677
11 RandomForestRegressor 0.4779574483287953 13.056004900000062
12 ExtraTreesRegressor 0.4994834890638471 1.7028706000000966
13 BaggingRegressor 0.41813406357742 1.2051542999997764
14 GradientBoostingRegressor 0.3223761613299607 4.173846099999992
15 AdaBoostRegressor 0.20700608833439227 0.6709999999998217
18 HistGradientBoostingRegressor 0.4239913847044323 0.7730876000000535
19 KNeighborsRegressor 0.18152755024686895 0.06760649999978341
20 RadiusNeighborsRegressor -0.0004093191012661812 0.708230199999889
21 MLPRegressor 0.19175110270945106 6.130972700000257
[22]:
import pandas
df = pandas.DataFrame(dict(model=names, r2=r2s, duree=durations))
df = df[["model", "r2", "duree"]]
df.sort_values("r2")
[22]:
| model | r2 | duree | |
|---|---|---|---|
| 4 | TheilSenRegressor | -0.294424 | 1.606173 |
| 2 | PassiveAggressiveRegressor | -0.062840 | 0.022322 |
| 16 | RadiusNeighborsRegressor | -0.000409 | 0.708230 |
| 8 | TweedieRegressor | -0.000127 | 0.061568 |
| 7 | GammaRegressor | -0.000113 | 0.029067 |
| 6 | PoissonRegressor | 0.001320 | 0.090246 |
| 3 | SGDRegressor | 0.008294 | 0.019756 |
| 5 | RANSACRegressor | 0.099806 | 0.259700 |
| 0 | HuberRegressor | 0.152649 | 1.074713 |
| 1 | LinearRegression | 0.179899 | 0.029094 |
| 15 | KNeighborsRegressor | 0.181528 | 0.067606 |
| 17 | MLPRegressor | 0.191751 | 6.130973 |
| 13 | AdaBoostRegressor | 0.207006 | 0.671000 |
| 12 | GradientBoostingRegressor | 0.322376 | 4.173846 |
| 11 | BaggingRegressor | 0.418134 | 1.205154 |
| 14 | HistGradientBoostingRegressor | 0.423991 | 0.773088 |
| 9 | RandomForestRegressor | 0.477957 | 13.056005 |
| 10 | ExtraTreesRegressor | 0.499483 | 1.702871 |
On filtre les valeurs inférieures à -1.
[23]:
df = df[df.r2 >= -1]
[24]:
import matplotlib.pyplot as plt
fig, ax = plt.subplots(figsize=(12, 4))
df.plot(x="duree", y="r2", kind="scatter", ax=ax)
for row in df.itertuples():
t, y, x = row[1:4]
if t[0] in {"B", "L"}:
ax.text(x, y, t, rotation=30, ha="left", va="bottom")
else:
ax.text(x, y, t, rotation=-40)
ax.set_title("Comparaison des modèles selon le temps d'apprentissage / r2");
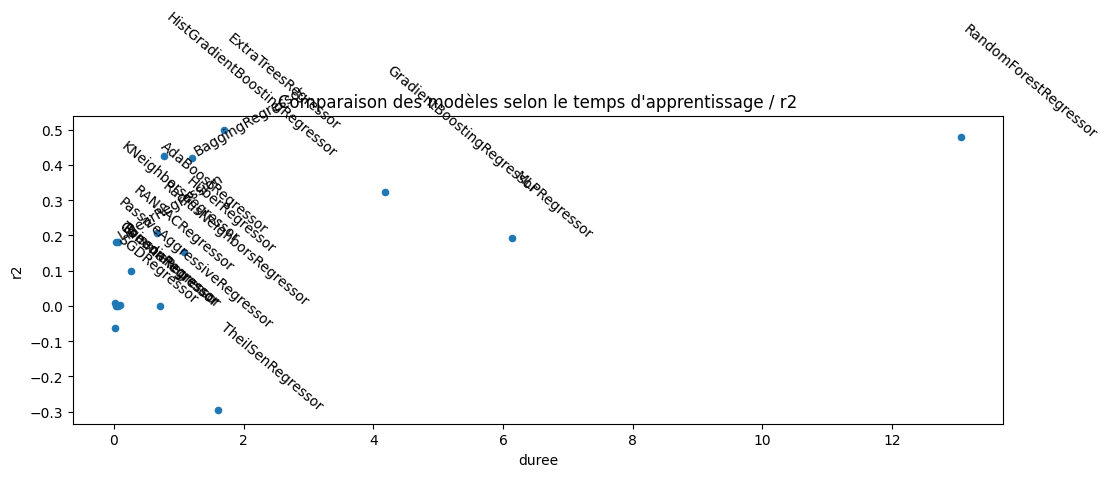
L’estimateur RANSACRegressor produit un très négatif. Regardons plus en détail.
[25]:
pred = regressors["RANSACRegressor"].predict(X_test_norm)
[26]:
import numpy.random
df = pandas.DataFrame(dict(pred=pred, expected=y_test))
df["expected"] += numpy.random.random(df.shape[0]) * 0.5
ax = df.plot(x="expected", y="pred", kind="scatter", figsize=(4, 4), linewidths=0.1)
ax.set_title("Graphe XY - valeurs attendues / prédites");
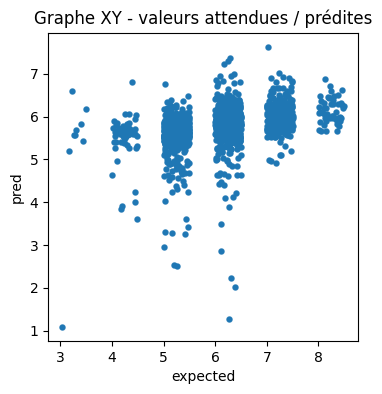
Essayons de voir avec la densité.
[30]:
import seaborn
ax = seaborn.jointplot(
df, x="expected", y="pred", kind="kde", size=4, space=0, ylim=(1, 9)
)
ax.ax_marg_y.set_title("Densité XY - valeur attendues / prédites");
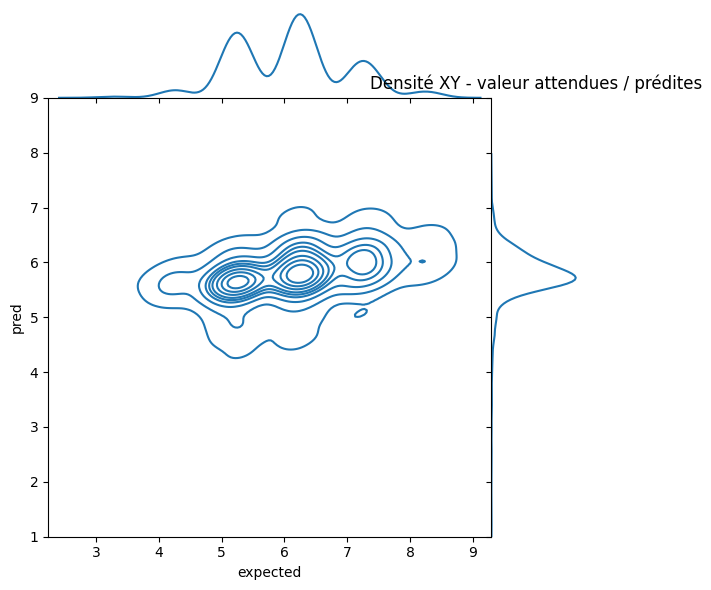
Pas facile à voir. Essayons de voir autrement en triant les prédictions et les valeurs attendues par ordre.
[31]:
sv = df.sort_values(["expected", "pred"]).reset_index(drop=True)
[32]:
import matplotlib.pyplot as plt
fig, ax = plt.subplots(figsize=(12, 3))
ax.plot(sv["pred"], label="RANSAC", lw=0.1)
ax.plot(list(sorted(df["expected"])), label="expected")
ax.plot(list(sorted(df["pred"])), label="RANSAC trié")
ax.set_ylim([0, 9])
ax.set_title("Valeur attendues et prédites triées par ordre croissant")
ax.legend();
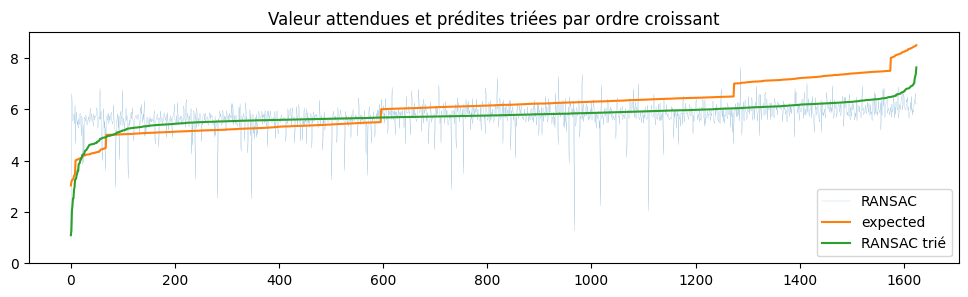
Le modèle est plutôt agité.
[33]:
regressors["RANSACRegressor"].estimator_.intercept_
[33]:
-0.3658125914901369
Pour s’assurer que les résultats sont fiables, il faut recommencer avec une validation croisée.
[34]:
import numpy.random
rnd = numpy.random.permutation(range(X_norm.shape[0]))
xns = X_norm[rnd, :]
yns = yn[rnd]
xns.shape, yns.shape
[34]:
((6497, 11), (6497,))
[35]:
from sklearn.model_selection import cross_val_score
def score_model_cross(xn, yn, model):
res = cross_val_score(model, xn, yn, cv=3)
return res.mean(), min(res), max(res)
score_model_cross(xns, yns, LinearRegression())
[35]:
(0.1799448611096674, 0.16609433123462292, 0.20072693380863527)
[41]:
from teachpyx.ext_test_case import unit_test_going
r2s = []
mis = []
mas = []
names = []
durations = []
regressors = {}
for i, model in enumerate(models):
if model.__name__ in {
"ARDRegression",
"VotingRegressor",
"QuantileRegressor",
"MLPRegressor",
"RandomForestRegressor",
"GradientBoostingRegressor", # too long
}:
continue
if unit_test_going() and not model.__name__ not in {
"LinearRegression",
"SGDRegressor",
}:
continue
try:
reg = model()
except Exception as e:
print("Skip", model)
continue
begin = perf_counter()
r2, mi, ma = score_model_cross(xns, yns, reg)
duree = perf_counter() - begin
r2s.append(r2)
mis.append(mi)
mas.append(ma)
names.append(model.__name__)
durations.append(duree)
regressors[model.__name__] = reg
print(i, model.__name__, r2, mi, ma, "t=%r" % duree)
1 HuberRegressor 0.16445120603440527 0.15765403941480316 0.1786143072062636 t=1.435193799999979
2 LinearRegression 0.1799448611096674 0.16609433123462292 0.20072693380863527 t=0.047846299999946496
3 PassiveAggressiveRegressor -0.8998543024559884 -2.5295972496836714 -0.08733290068040933 t=0.13489810000010038
5 SGDRegressor 0.00973904451751757 -0.004417086935930126 0.01816079178339436 t=0.11661329999969894
6 TheilSenRegressor -0.3139016667645712 -0.41907116748283557 -0.224530428533247 t=6.356290799999897
7 RANSACRegressor -5.4384892594797405 -12.400989908758083 -1.1372714492625668 t=1.0156056000000717
8 PoissonRegressor 0.0011212946218149167 -0.00022109795646318986 0.0020054507493099116 t=0.3450553999996373
9 GammaRegressor -0.0005558618017731165 -0.0021320248191363245 0.00030404381558213345 t=0.25818170000002283
10 TweedieRegressor -0.0005670558820739524 -0.002194268482211603 0.0003060535183355695 t=0.3232327999999143
12 ExtraTreesRegressor 0.5150920905590446 0.48820834539223157 0.5338810535848475 t=9.26701920000005
13 BaggingRegressor 0.43686266293982196 0.401753167463089 0.4758920210408679 t=7.2353933999997935
15 AdaBoostRegressor 0.18291148330046114 0.15479217393907074 0.22140473381847336 t=3.640226099999836
Skip <class 'sklearn.ensemble._stacking.StackingRegressor'>
18 HistGradientBoostingRegressor 0.41442261067915653 0.3888441812635889 0.43299359267487436 t=3.9275103999998464
19 KNeighborsRegressor 0.1653286657553436 0.0875423648134045 0.19788204011638 t=0.1673464000000422
20 RadiusNeighborsRegressor -0.0008592314579911386 -0.002575734438727606 0.00030815848198539886 t=3.3217153999999027
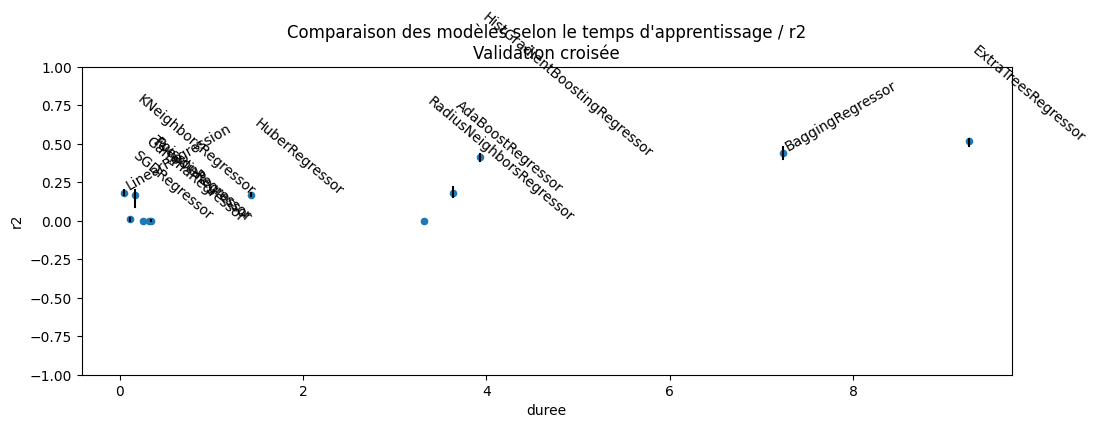
[44]:
df = pandas.DataFrame(dict(model=names, r2=r2s, min=mis, max=mas, duree=durations))
df = df[["model", "r2", "min", "max", "duree"]]
df.sort_values("r2")
[44]:
| model | r2 | min | max | duree | |
|---|---|---|---|---|---|
| 5 | RANSACRegressor | -5.438489 | -12.400990 | -1.137271 | 1.015606 |
| 2 | PassiveAggressiveRegressor | -0.899854 | -2.529597 | -0.087333 | 0.134898 |
| 4 | TheilSenRegressor | -0.313902 | -0.419071 | -0.224530 | 6.356291 |
| 14 | RadiusNeighborsRegressor | -0.000859 | -0.002576 | 0.000308 | 3.321715 |
| 8 | TweedieRegressor | -0.000567 | -0.002194 | 0.000306 | 0.323233 |
| 7 | GammaRegressor | -0.000556 | -0.002132 | 0.000304 | 0.258182 |
| 6 | PoissonRegressor | 0.001121 | -0.000221 | 0.002005 | 0.345055 |
| 3 | SGDRegressor | 0.009739 | -0.004417 | 0.018161 | 0.116613 |
| 0 | HuberRegressor | 0.164451 | 0.157654 | 0.178614 | 1.435194 |
| 13 | KNeighborsRegressor | 0.165329 | 0.087542 | 0.197882 | 0.167346 |
| 1 | LinearRegression | 0.179945 | 0.166094 | 0.200727 | 0.047846 |
| 11 | AdaBoostRegressor | 0.182911 | 0.154792 | 0.221405 | 3.640226 |
| 12 | HistGradientBoostingRegressor | 0.414423 | 0.388844 | 0.432994 | 3.927510 |
| 10 | BaggingRegressor | 0.436863 | 0.401753 | 0.475892 | 7.235393 |
| 9 | ExtraTreesRegressor | 0.515092 | 0.488208 | 0.533881 | 9.267019 |
[45]:
import matplotlib.pyplot as plt
fig, ax = plt.subplots(figsize=(12, 4))
df[df["min"] > -0.1].plot(x="duree", y="r2", kind="scatter", ax=ax)
for row in df.itertuples():
t, y, mi, ma, x = row[1:6]
if mi < -0.1:
continue
ax.plot([x, x], [mi, ma], color="black")
if t[0] in {"B", "L"}:
ax.text(x, y, t, rotation=30, ha="left", va="bottom")
else:
ax.text(x, y, t, rotation=-40)
ax.set_ylim([-1, 1])
ax.set_title(
"Comparaison des modèles selon le temps d'apprentissage / r2\nValidation croisée"
);
Le modèle RANSAC est conçu pour apprendre un modèle linéaire et réduire l’ensemble d’apprentissage aux points non aberrants. Dans notre cas, vu le peu d’exemples pour les notes élevées, il est très probable que celles-ci disparaissent des observations choisies pour estimer le modèle : le modèle choisit d’évincer les exemples pour lesquels l’erreur est la plus grande en considérant que cela est une indication du fait qu’ils sont aberrants. Malgré cela, sa faible capacité à prévoir vient du fait que la majorité des vins ont une note entre 4 et 6 et que finalement, il y a peu de différences : ils sont tous moyens et la différence s’explique par d’autres facteurs comme le juge ayant donné la note.