Régression logistique en 2D¶
Prédire la couleur d’un vin à partir de ses composants.
[1]:
%matplotlib inline
[4]:
from teachpyx.datasets import load_wines_dataset
data = load_wines_dataset()
X = data.drop(["quality", "color"], axis=1)
y = data["color"]
[5]:
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y)
[6]:
from statsmodels.discrete.discrete_model import Logit
model = Logit(y_train == "white", X_train)
res = model.fit()
Optimization terminated successfully.
Current function value: 0.048414
Iterations 11
[7]:
res.summary2()
[7]:
| Model: | Logit | Method: | MLE |
| Dependent Variable: | color | Pseudo R-squared: | 0.913 |
| Date: | 2024-01-23 00:52 | AIC: | 493.7476 |
| No. Observations: | 4872 | BIC: | 565.1515 |
| Df Model: | 10 | Log-Likelihood: | -235.87 |
| Df Residuals: | 4861 | LL-Null: | -2717.5 |
| Converged: | 1.0000 | LLR p-value: | 0.0000 |
| No. Iterations: | 11.0000 | Scale: | 1.0000 |
| Coef. | Std.Err. | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| fixed_acidity | -1.4541 | 0.1515 | -9.5981 | 0.0000 | -1.7511 | -1.1572 |
| volatile_acidity | -11.3716 | 0.9995 | -11.3771 | 0.0000 | -13.3306 | -9.4126 |
| citric_acid | 1.7492 | 1.1280 | 1.5507 | 0.1210 | -0.4616 | 3.9599 |
| residual_sugar | 0.1246 | 0.0600 | 2.0756 | 0.0379 | 0.0069 | 0.2422 |
| chlorides | -32.7390 | 3.9560 | -8.2758 | 0.0000 | -40.4926 | -24.9854 |
| free_sulfur_dioxide | -0.0505 | 0.0134 | -3.7724 | 0.0002 | -0.0768 | -0.0243 |
| total_sulfur_dioxide | 0.0632 | 0.0050 | 12.6896 | 0.0000 | 0.0534 | 0.0730 |
| density | 42.0110 | 4.2093 | 9.9806 | 0.0000 | 33.7610 | 50.2610 |
| pH | -8.7417 | 0.9800 | -8.9204 | 0.0000 | -10.6624 | -6.8210 |
| sulphates | -8.8918 | 1.0237 | -8.6857 | 0.0000 | -10.8983 | -6.8853 |
| alcohol | 0.4150 | 0.1233 | 3.3656 | 0.0008 | 0.1733 | 0.6567 |
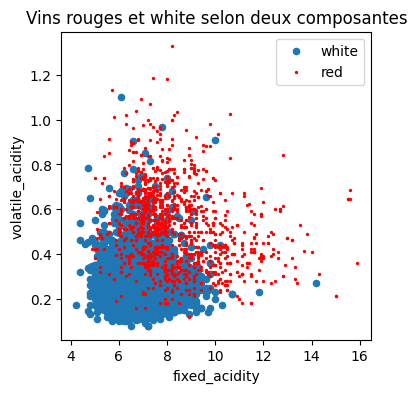
On ne garde que les deux premières.
[8]:
X_train2 = X_train.iloc[:, :2]
[9]:
import pandas
df = pandas.DataFrame(X_train2.copy())
df["y"] = y_train
import matplotlib.pyplot as plt
fig, ax = plt.subplots(1, 1, figsize=(4, 4))
df[df.y == "white"].plot(
x="fixed_acidity", y="volatile_acidity", ax=ax, kind="scatter", label="white"
)
df[df.y == "red"].plot(
x="fixed_acidity",
y="volatile_acidity",
ax=ax,
kind="scatter",
label="red",
color="red",
s=2,
)
ax.set_title("Vins rouges et white selon deux composantes");
[10]:
from sklearn.linear_model import LogisticRegression
model = LogisticRegression()
model.fit(X_train2, y_train == "white")
[10]:
LogisticRegression()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
LogisticRegression()
[11]:
model.coef_, model.intercept_
[11]:
(array([[ -1.11120776, -11.79383309]]), array([13.83313405]))
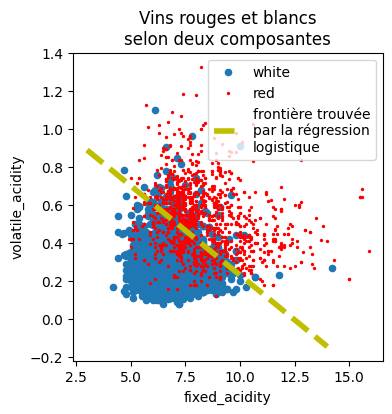
On trace cette droite sur le graphique.
[12]:
x0 = 3
y0 = -(model.coef_[0, 0] * x0 + model.intercept_) / model.coef_[0, 1]
x1 = 14
y1 = -(model.coef_[0, 0] * x1 + model.intercept_) / model.coef_[0, 1]
x0, y0, x1, y1
[12]:
(3, array([0.89025431]), 14, array([-0.14615898]))
[13]:
import matplotlib.pyplot as plt
fig, ax = plt.subplots(1, 1, figsize=(4, 4))
df[df.y == "white"].plot(
x="fixed_acidity", y="volatile_acidity", ax=ax, kind="scatter", label="white"
)
df[df.y == "red"].plot(
x="fixed_acidity",
y="volatile_acidity",
ax=ax,
kind="scatter",
label="red",
color="red",
s=2,
)
ax.plot(
[x0, x1],
[y0, y1],
"y--",
lw=4,
label="frontière trouvée\npar la régression\nlogistique",
)
ax.legend()
ax.set_title("Vins rouges et blancs\nselon deux composantes");