Factorisation et matrice et ACP¶
Un exemple pour montrer l’équivalence entre l’ACP et une factorisation de matrice.
[2]:
%matplotlib inline
Factorisation de matrices¶
[3]:
def erreur_mf(M, W, H):
d = M - W @ H
a = d.ravel()
e = a @ a.T
e**0.5 / (M.shape[0] * M.shape[1])
return e
On crée un nuage de points avec que des coordonnées positives pour satisfaire les hypothèses de la factorisation de matrices.
[4]:
from numpy.random import rand
M = rand(2, 20)
M[1, :] += 3 * M[0, :]
M
[4]:
array([[ 0.81960047, 0.63887134, 0.74019269, 0.96110175, 0.0685406 ,
0.11103301, 0.06033529, 0.67913157, 0.10460611, 0.98860048,
0.50497448, 0.26893866, 0.73143267, 0.32617974, 0.1332449 ,
0.83328515, 0.3775355 , 0.69163261, 0.53095348, 0.15601268],
[ 2.48031078, 2.2279066 , 2.85929872, 3.27833973, 0.27323095,
0.53806662, 0.48019992, 2.09428487, 0.40521666, 3.94539474,
2.36639105, 1.66857684, 3.14027534, 1.94032092, 1.22602705,
3.09679803, 1.696636 , 2.69144798, 1.84350664, 1.16862532]])
[5]:
from sklearn.decomposition import NMF
mf = NMF(1)
W = mf.fit_transform(M)
H = mf.components_
erreur_mf(M, W, H)
[5]:
0.19729615330190822
[6]:
wh = W @ H
[7]:
import matplotlib.pyplot as plt
fig, ax = plt.subplots(1, 1)
ax.plot(M[0, :], M[1, :], "ob")
ax.plot(wh[0, :], wh[1, :], "or")
ax.set_xlim([0, 1])
ax.set_ylim([0, 4])
[7]:
(0, 4)
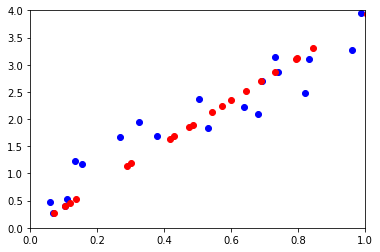
ACP : analyse en composantes principales¶
[8]:
from sklearn.decomposition import PCA
pca = PCA(n_components=1)
pca.fit(M.T)
[8]:
PCA(copy=True, iterated_power='auto', n_components=1, random_state=None,
svd_solver='auto', tol=0.0, whiten=False)
[9]:
projected_points = pca.inverse_transform(pca.transform(M.T))
pj = projected_points.T
[10]:
import matplotlib.pyplot as plt
fig, ax = plt.subplots(1, 1)
ax.plot(M[0, :], M[1, :], "ob")
ax.plot(wh[0, :], wh[1, :], "or")
ax.plot(pj[0, :], pj[1, :], "og")
ax.set_xlim([0, 1])
ax.set_ylim([0, 4])
[10]:
(0, 4)
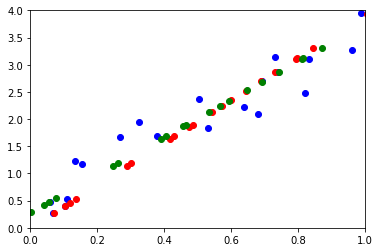
Les résultats ne sont pas exactement identiques car l”ACP centre le nuage de points par défaut. On utilise celui de statsmodels pour éviter cela.
[11]:
from statsmodels.multivariate.pca import PCA
pca = PCA(M.T, ncomp=1, standardize=False, demean=False, normalize=False)
pca
[11]:
Principal Component Analysis(nobs: 20, nvar: 2, transformation: None, normalization: False, number of components: 1, SVD, id: 0x1c01a2861d0)
[12]:
pj2 = pca.projection.T
[13]:
import matplotlib.pyplot as plt
fig, ax = plt.subplots(1, 1)
ax.plot(M[0, :], M[1, :], "ob")
# ax.plot(wh[0,:], wh[1,:], "or")
ax.plot(pj[0, :], pj[1, :], "og")
ax.plot(pj2[0, :], pj2[1, :], "oy")
ax.set_xlim([0, 1])
ax.set_ylim([0, 4])
[13]:
(0, 4)
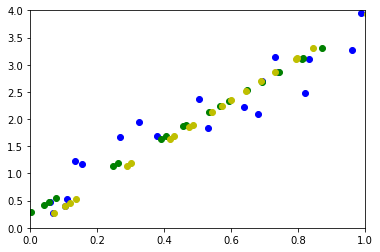
On retrouve exactement les mêmes résultats.
[14]: