Note
Go to the end to download the full example code.
Measuring CUDA performance with a vector sum¶
The objective is to measure the summation of all elements from a tensor.
nsys profile python _doc/examples/plot_bench_cuda_vector_sum.py
Vector Add¶
from tqdm import tqdm
import numpy
import matplotlib.pyplot as plt
from pandas import DataFrame
from teachcompute.ext_test_case import measure_time, unit_test_going
import torch
has_cuda = torch.cuda.is_available()
try:
from teachcompute.validation.cuda.cuda_example_py import (
vector_sum0,
vector_sum_atomic,
vector_sum6,
)
except ImportError:
has_cuda = False
def wrap_cuda_call(f, values):
torch.cuda.nvtx.range_push(f"CUDA f={f.__name__} dim={values.size}")
res = f(values)
torch.cuda.nvtx.range_pop()
return res
obs = []
dims = [2**10, 2**15, 2**20, 2**25, 2**28]
if unit_test_going():
dims = [10, 20, 30]
for dim in tqdm(dims):
values = numpy.ones((dim,), dtype=numpy.float32).ravel()
if has_cuda:
for f in [vector_sum0, vector_sum_atomic, vector_sum6]:
if f == vector_sum_atomic and dim > 2**20:
continue
diff = numpy.abs(wrap_cuda_call(f, values) - (values.sum()))
res = measure_time(
lambda f=f, values=values: wrap_cuda_call(f, values), max_time=0.5
)
obs.append(
dict(
dim=dim,
size=values.size,
time=res["average"],
fct=f"CUDA-{f.__name__}",
time_per_element=res["average"] / dim,
diff=diff,
)
)
diff = 0
res = measure_time(lambda values=values: values.sum(), max_time=0.5)
obs.append(
dict(
dim=dim,
size=values.size,
time=res["average"],
fct="numpy",
time_per_element=res["average"] / dim,
diff=0,
)
)
df = DataFrame(obs)
piv = df.pivot(index="dim", columns="fct", values="time_per_element")
print(piv)
0%| | 0/5 [00:00<?, ?it/s]
20%|██ | 1/5 [00:02<00:09, 2.46s/it]
40%|████ | 2/5 [00:04<00:06, 2.33s/it]
60%|██████ | 3/5 [00:08<00:06, 3.07s/it]
80%|████████ | 4/5 [00:10<00:02, 2.66s/it]
100%|██████████| 5/5 [00:15<00:00, 3.46s/it]
100%|██████████| 5/5 [00:15<00:00, 3.11s/it]
fct CUDA-vector_sum0 CUDA-vector_sum6 CUDA-vector_sum_atomic numpy
dim
1024 2.274043e-06 2.272027e-06 3.184170e-06 1.802920e-09
32768 8.648022e-08 8.932439e-08 5.989448e-07 3.572639e-10
1048576 4.994521e-09 3.494095e-09 3.595154e-07 3.355802e-10
33554432 1.028890e-09 9.966636e-10 NaN 4.411365e-10
268435456 1.354863e-09 9.829053e-10 NaN 4.245788e-10
Plots¶
piv_diff = df.pivot(index="dim", columns="fct", values="diff")
piv_time = df.pivot(index="dim", columns="fct", values="time")
fig, ax = plt.subplots(1, 3, figsize=(12, 6))
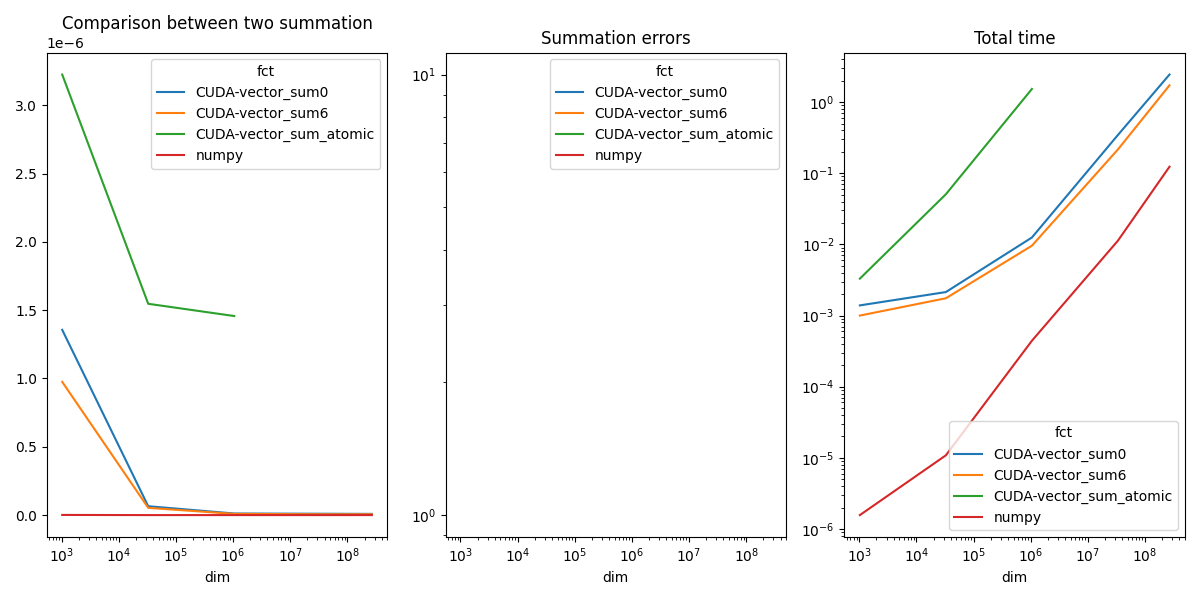
piv.plot(ax=ax[0], logx=True, title="Comparison between two summation")
piv_diff.plot(ax=ax[1], logx=True, logy=True, title="Summation errors")
piv_time.plot(ax=ax[2], logx=True, logy=True, title="Total time")
fig.tight_layout()
fig.savefig("plot_bench_cuda_vector_sum.png")
~/vv/this312/lib/python3.12/site-packages/pandas/plotting/_matplotlib/core.py:822: UserWarning: Data has no positive values, and therefore cannot be log-scaled.
labels = axis.get_majorticklabels() + axis.get_minorticklabels()
CUDA seems very slow but in fact, all the time is spent in moving the data from the CPU memory (Host) to the GPU memory (device).
Total running time of the script: (0 minutes 17.094 seconds)