Note
Go to the end to download the full example code.
Measuring CPU performance with a parallelized vector sum¶
The example compares the time spend in computing the sum of all coefficients of a matrix when the function walks through the coefficients by rows or by columns when the computation is parallelized.
Vector Sum¶
from tqdm import tqdm
import numpy
import matplotlib.pyplot as plt
from pandas import DataFrame
from teachcompute.ext_test_case import measure_time, unit_test_going
from teachcompute.validation.cpu._validation import (
vector_sum_array as vector_sum,
vector_sum_array_parallel as vector_sum_parallel,
)
obs = []
dims = [500, 700, 800, 900, 1000, 1100, 1200, 1300, 1400, 1500, 1600, 1700, 1800, 2000]
if unit_test_going():
dims = [10, 20]
for dim in tqdm(dims):
values = numpy.ones((dim, dim), dtype=numpy.float32).ravel()
diff = abs(vector_sum(dim, values, True) - dim**2)
res = measure_time(
lambda dim=dim, values=values: vector_sum(dim, values, True), max_time=0.5
)
obs.append(
dict(
dim=dim,
size=values.size,
time=res["average"],
direction="rows",
time_per_element=res["average"] / dim**2,
diff=diff,
)
)
res = measure_time(
lambda dim=dim, values=values: vector_sum_parallel(dim, values, True),
max_time=0.5,
)
obs.append(
dict(
dim=dim,
size=values.size,
time=res["average"],
direction="rows//",
time_per_element=res["average"] / dim**2,
diff=diff,
)
)
diff = abs(vector_sum(dim, values, False) - dim**2)
res = measure_time(
lambda dim=dim, values=values: vector_sum_parallel(dim, values, False),
max_time=0.5,
)
obs.append(
dict(
dim=dim,
size=values.size,
time=res["average"],
direction="cols//",
time_per_element=res["average"] / dim**2,
diff=diff,
)
)
df = DataFrame(obs)
piv = df.pivot(index="dim", columns="direction", values="time_per_element")
print(piv)
0%| | 0/14 [00:00<?, ?it/s]
7%|▋ | 1/14 [00:01<00:23, 1.82s/it]
14%|█▍ | 2/14 [00:03<00:21, 1.78s/it]
21%|██▏ | 3/14 [00:05<00:19, 1.79s/it]
29%|██▊ | 4/14 [00:07<00:18, 1.88s/it]
36%|███▌ | 5/14 [00:09<00:16, 1.80s/it]
43%|████▎ | 6/14 [00:10<00:14, 1.79s/it]
50%|█████ | 7/14 [00:12<00:12, 1.79s/it]
57%|█████▋ | 8/14 [00:14<00:10, 1.80s/it]
64%|██████▍ | 9/14 [00:16<00:09, 1.80s/it]
71%|███████▏ | 10/14 [00:17<00:06, 1.75s/it]
79%|███████▊ | 11/14 [00:19<00:05, 1.72s/it]
86%|████████▌ | 12/14 [00:21<00:03, 1.72s/it]
93%|█████████▎| 13/14 [00:23<00:01, 1.74s/it]
100%|██████████| 14/14 [00:24<00:00, 1.75s/it]
100%|██████████| 14/14 [00:24<00:00, 1.77s/it]
direction cols// rows rows//
dim
500 2.274721e-10 8.563124e-10 2.139315e-10
700 3.367411e-10 8.591378e-10 2.029837e-10
800 2.705322e-10 8.963944e-10 2.034740e-10
900 2.630441e-10 9.021412e-10 2.027335e-10
1000 3.515007e-10 8.520763e-10 2.201897e-10
1100 5.244538e-10 8.966395e-10 2.417822e-10
1200 7.161109e-10 9.007439e-10 2.046541e-10
1300 1.064342e-09 9.320889e-10 2.018137e-10
1400 1.174268e-09 9.464296e-10 2.017736e-10
1500 1.395570e-09 9.077340e-10 1.896674e-10
1600 1.496557e-09 8.895338e-10 1.838071e-10
1700 1.443899e-09 9.582795e-10 1.983384e-10
1800 1.663758e-09 9.282506e-10 1.834979e-10
2000 1.594137e-09 9.280747e-10 1.865809e-10
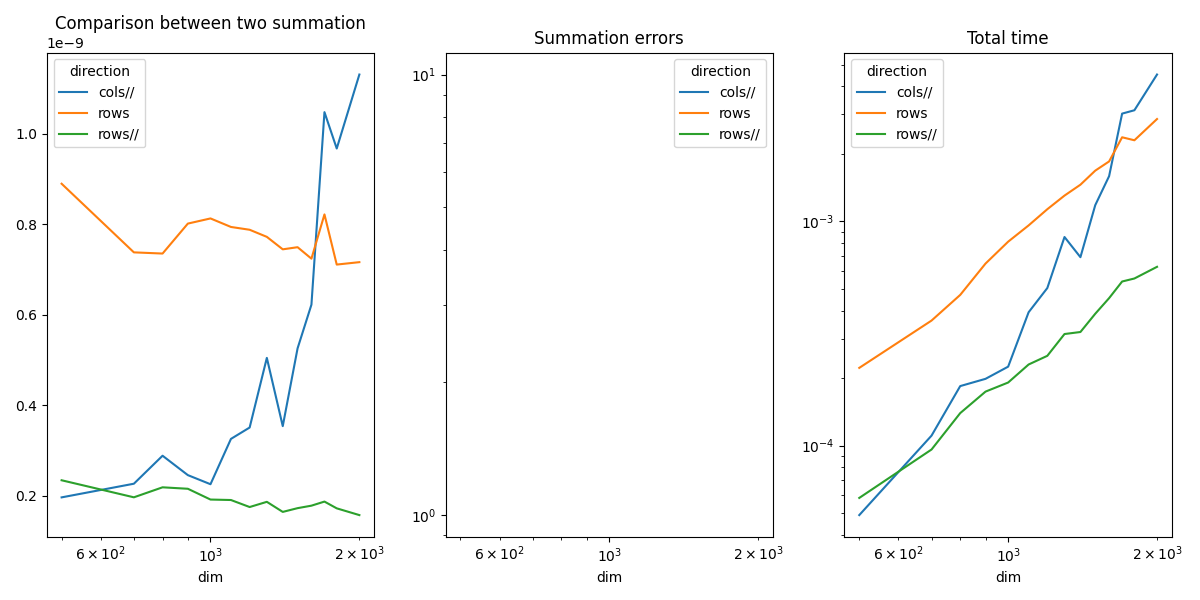
Plots¶
piv_diff = df.pivot(index="dim", columns="direction", values="diff")
piv_time = df.pivot(index="dim", columns="direction", values="time")
fig, ax = plt.subplots(1, 3, figsize=(12, 6))
piv.plot(ax=ax[0], logx=True, title="Comparison between two summation")
piv_diff.plot(ax=ax[1], logx=True, logy=True, title="Summation errors")
piv_time.plot(ax=ax[2], logx=True, logy=True, title="Total time")
fig.tight_layout()
fig.savefig("plot_bench_cpu_vector_sum_parallel.png")
~/vv/this312/lib/python3.12/site-packages/pandas/plotting/_matplotlib/core.py:822: UserWarning: Data has no positive values, and therefore cannot be log-scaled.
labels = axis.get_majorticklabels() + axis.get_minorticklabels()
The summation by rows is much faster as expected. That explains why it is usually more efficient to transpose the first matrix before a matrix multiplication. Parallelization is faster.
Total running time of the script: (0 minutes 25.824 seconds)