Note
Go to the end to download the full example code.
Regression with confidence interval¶
The notebook computes confidence intervals with bootstrapping and quantile regression on a simple problem.
Some data¶
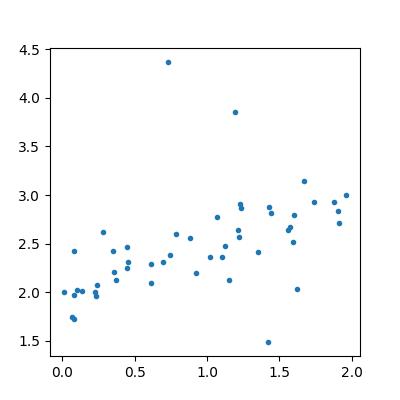
The data follows the formula:
. Noises
follows the laws
,
,
. The second part of the noise
adds some bigger noise but not always.
import numpy
from numpy.random import binomial, rand, randn
import pandas
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.gaussian_process import GaussianProcessRegressor
from sklearn.gaussian_process.kernels import (
RBF,
ConstantKernel as C,
WhiteKernel,
)
from sklearn.linear_model import LinearRegression
from sklearn.tree import DecisionTreeRegressor
from mlinsights.mlmodel import IntervalRegressor, QuantileLinearRegression
N = 200
X = rand(N, 1) * 2
eps = randn(N, 1) * 0.2
eps2 = randn(N, 1) + 1
bin = binomial(2, 0.05, size=(N, 1))
y = (0.5 * X + eps + 2 + eps2 * bin).ravel()
[<matplotlib.lines.Line2D object at 0x79fc064f76e0>]
Confidence interval with a linear regression¶
# The object fits many times the same learner, every training is done on a
# resampling of the training dataset.
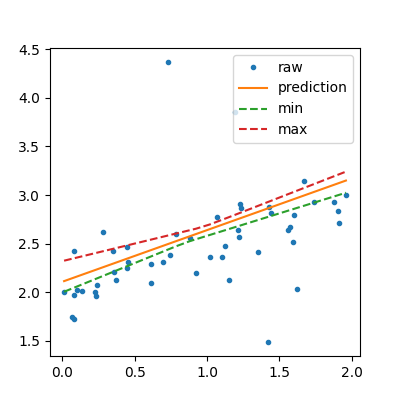
lin = IntervalRegressor(LinearRegression())
lin.fit(X_train, y_train)
sorted_X = numpy.array(list(sorted(X_test)))
pred = lin.predict(sorted_X)
bootstrapped_pred = lin.predict_sorted(sorted_X)
min_pred = bootstrapped_pred[:, 0]
max_pred = bootstrapped_pred[:, bootstrapped_pred.shape[1] - 1]
<matplotlib.legend.Legend object at 0x79fc00608290>
Higher confidence interval¶
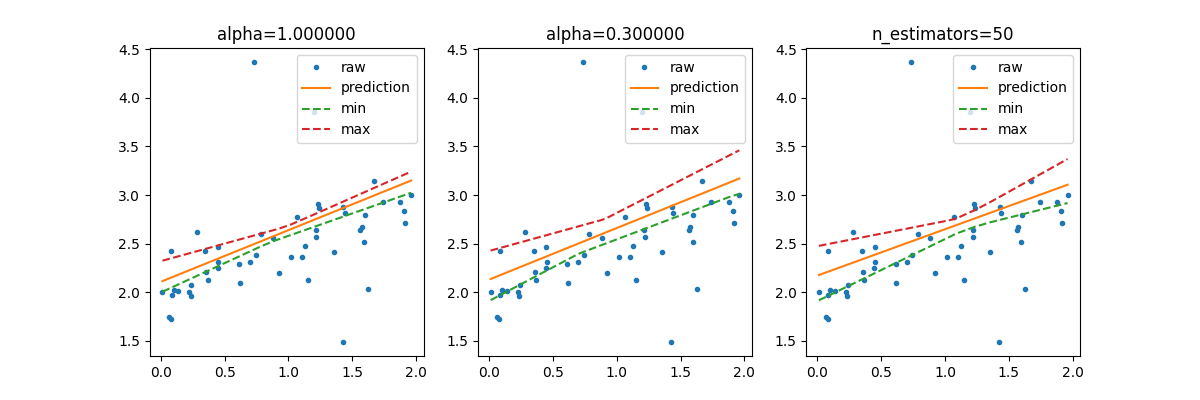
# It is possible to use smaller resample of the training dataset or we can
# increase the number of resamplings.
lin2 = IntervalRegressor(LinearRegression(), alpha=0.3)
lin2.fit(X_train, y_train)
lin3 = IntervalRegressor(LinearRegression(), n_estimators=50)
lin3.fit(X_train, y_train)
pred2 = lin2.predict(sorted_X)
bootstrapped_pred2 = lin2.predict_sorted(sorted_X)
min_pred2 = bootstrapped_pred2[:, 0]
max_pred2 = bootstrapped_pred2[:, bootstrapped_pred2.shape[1] - 1]
pred3 = lin3.predict(sorted_X)
bootstrapped_pred3 = lin3.predict_sorted(sorted_X)
min_pred3 = bootstrapped_pred3[:, 0]
max_pred3 = bootstrapped_pred3[:, bootstrapped_pred3.shape[1] - 1]
fig, ax = plt.subplots(1, 3, figsize=(12, 4))
ax[0].plot(X_test, y_test, ".", label="raw")
ax[0].plot(sorted_X, pred, label="prediction")
ax[0].plot(sorted_X, min_pred, "--", label="min")
ax[0].plot(sorted_X, max_pred, "--", label="max")
ax[0].legend()
ax[0].set_title("alpha=%f" % lin.alpha)
ax[1].plot(X_test, y_test, ".", label="raw")
ax[1].plot(sorted_X, pred2, label="prediction")
ax[1].plot(sorted_X, min_pred2, "--", label="min")
ax[1].plot(sorted_X, max_pred2, "--", label="max")
ax[1].set_title("alpha=%f" % lin2.alpha)
ax[1].legend()
ax[2].plot(X_test, y_test, ".", label="raw")
ax[2].plot(sorted_X, pred3, label="prediction")
ax[2].plot(sorted_X, min_pred3, "--", label="min")
ax[2].plot(sorted_X, max_pred3, "--", label="max")
ax[2].set_title("n_estimators=%d" % lin3.n_estimators)
ax[2].legend()
<matplotlib.legend.Legend object at 0x79fc07c85d00>
With decision trees¶
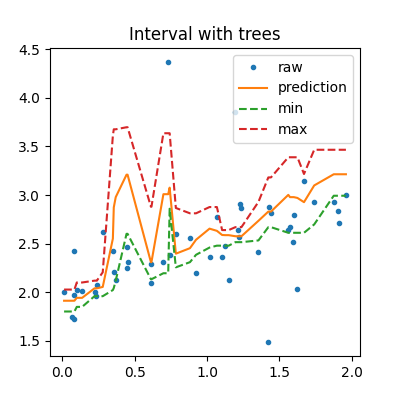
tree = IntervalRegressor(DecisionTreeRegressor(min_samples_leaf=10))
tree.fit(X_train, y_train)
pred_tree = tree.predict(sorted_X)
b_pred_tree = tree.predict_sorted(sorted_X)
min_pred_tree = b_pred_tree[:, 0]
max_pred_tree = b_pred_tree[:, b_pred_tree.shape[1] - 1]
<matplotlib.legend.Legend object at 0x79fc074b0290>
In that case, the prediction is very similar to the one a random forest would produce as it is an average of the predictions made by 10 trees.
Regression quantile¶
The last way tries to fit two regressions for quantiles 0.05 and 0.95.
m = QuantileLinearRegression()
q1 = QuantileLinearRegression(quantile=0.05)
q2 = QuantileLinearRegression(quantile=0.95)
for model in [m, q1, q2]:
model.fit(X_train, y_train)
[<matplotlib.lines.Line2D object at 0x79fb08544350>]
<matplotlib.legend.Legend object at 0x79fb09ef4770>
With a non linear model… but the model QuantileMLPRegressor only implements the regression with quantile 0.5.
With seaborn¶
It uses a theoritical way to compute the confidence interval by computing the confidence interval on the parameters first.
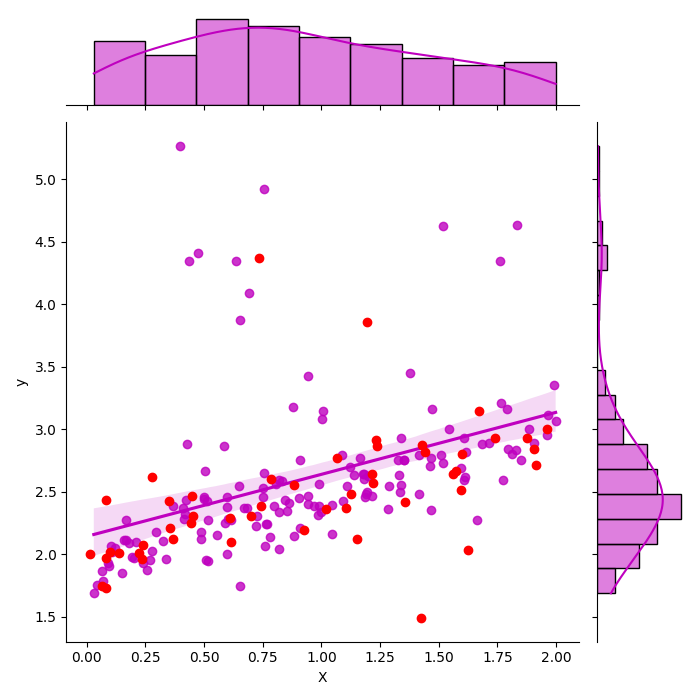
[<matplotlib.lines.Line2D object at 0x79fc0646b230>]
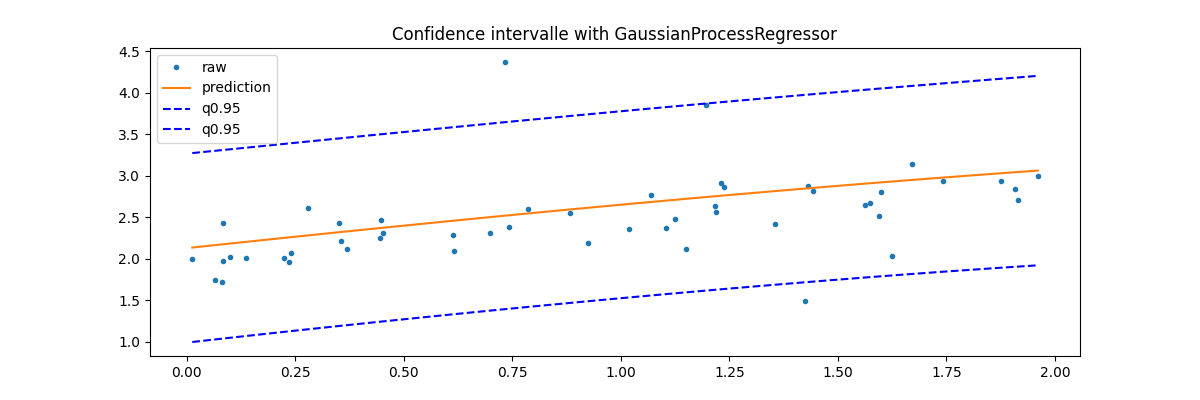
GaussianProcessRegressor¶
Last option with this example Gaussian Processes regression: basic introductory example which computes the standard deviation for every prediction. It can then be used to show an interval confidence.
y_pred, sigma = gp.predict(sorted_X, return_std=True)
fig, ax = plt.subplots(1, 1, figsize=(12, 4))
ax.plot(X_test, y_test, ".", label="raw")
ax.plot(sorted_X, y_pred, label="prediction")
ax.plot(sorted_X, y_pred + sigma * 1.96, "b--", label="q0.95")
ax.plot(sorted_X, y_pred - sigma * 1.96, "b--", label="q0.95")
ax.set_title("Confidence intervalle with GaussianProcessRegressor")
ax.legend()
<matplotlib.legend.Legend object at 0x79fc0647bdd0>
Total running time of the script: (0 minutes 2.094 seconds)