command lines#
bench#
The function compares the execution of a random forest regressor with different implementation of the tree implementations. Example:
python -m onnx_extended bench -r 3 -f 100 -d 12 -v -t 200 -b 10000 -n 100 \
-e onnxruntime,CReferenceEvaluator,onnxruntime-customops
Output example:
[bench_trees] 11:24:56.852193 create tree
[bench_trees] 11:24:59.094062 create forest with 200 trees
[bench_trees] 11:25:00.593993 modelsize 30961.328 Kb
[bench_trees] 11:25:00.660829 create datasets
[bench_trees] 11:25:00.724519 create engines
[bench_trees] 11:25:00.724649 create engine 'onnxruntime'
[bench_trees] 11:25:05.237548 create engine 'CReferenceEvaluator'
[bench_trees] 11:25:08.616215 create engine 'onnxruntime-customops'
[bench_trees] 11:25:21.837973 benchmark
[bench_trees] 11:25:21.838058 test 'onnxruntime' warmup...
[bench_trees] 11:25:21.997798 test 'onnxruntime' benchmark...
[bench_trees] 11:25:28.298222 test 'onnxruntime' duration=0.06300305700002355
[bench_trees] 11:25:28.298340 test 'CReferenceEvaluator' warmup...
[bench_trees] 11:25:39.017608 test 'CReferenceEvaluator' benchmark...
[bench_trees] 11:25:57.116259 test 'CReferenceEvaluator' duration=0.18098569900001166
[bench_trees] 11:25:57.116340 test 'onnxruntime-customops' warmup...
[bench_trees] 11:25:57.276264 test 'onnxruntime-customops' benchmark...
[bench_trees] 11:26:03.638203 test 'onnxruntime-customops' duration=0.06361832300000969
[bench_trees] 11:26:03.638315 test 'onnxruntime' warmup...
[bench_trees] 11:26:03.793355 test 'onnxruntime' benchmark...
[bench_trees] 11:26:10.111718 test 'onnxruntime' duration=0.0631821729999865
[bench_trees] 11:26:10.111833 test 'CReferenceEvaluator' warmup...
[bench_trees] 11:26:10.493677 test 'CReferenceEvaluator' benchmark...
[bench_trees] 11:26:28.573696 test 'CReferenceEvaluator' duration=0.1807989760000055
[bench_trees] 11:26:28.573834 test 'onnxruntime-customops' warmup...
[bench_trees] 11:26:28.737896 test 'onnxruntime-customops' benchmark...
[bench_trees] 11:26:35.095252 test 'onnxruntime-customops' duration=0.06357246399998985
[bench_trees] 11:26:35.095367 test 'onnxruntime' warmup...
[bench_trees] 11:26:35.238863 test 'onnxruntime' benchmark...
[bench_trees] 11:26:41.230780 test 'onnxruntime' duration=0.05991804699999193
[bench_trees] 11:26:41.230903 test 'CReferenceEvaluator' warmup...
[bench_trees] 11:26:41.621822 test 'CReferenceEvaluator' benchmark...
[bench_trees] 11:26:59.714731 test 'CReferenceEvaluator' duration=0.180928322999971
[bench_trees] 11:26:59.714814 test 'onnxruntime-customops' warmup...
[bench_trees] 11:26:59.871232 test 'onnxruntime-customops' benchmark...
[bench_trees] 11:27:06.267876 test 'onnxruntime-customops' duration=0.06396529300000112
name repeat duration n_estimators number n_features max_depth batch_size
0 onnxruntime 0 0.063003 200 100 100 12 10000
1 CReferenceEvaluator 0 0.180986 200 100 100 12 10000
2 onnxruntime-customops 0 0.063618 200 100 100 12 10000
3 onnxruntime 1 0.063182 200 100 100 12 10000
4 CReferenceEvaluator 1 0.180799 200 100 100 12 10000
5 onnxruntime-customops 1 0.063572 200 100 100 12 10000
6 onnxruntime 2 0.059918 200 100 100 12 10000
7 CReferenceEvaluator 2 0.180928 200 100 100 12 10000
8 onnxruntime-customops 2 0.063965 200 100 100 12 10000
usage: bench [-h] [-d MAX_DEPTH] [-t N_ESTIMATORS] [-f N_FEATURES]
[-b BATCH_SIZE] [-n NUMBER] [-w WARMUP] [-r REPEAT] [-e ENGINE]
[-v] [-p] [-o OUTPUT]
Benchmarks kernel executions.
options:
-h, --help show this help message and exit
-d MAX_DEPTH, --max_depth MAX_DEPTH
max_depth for all trees
-t N_ESTIMATORS, --n_estimators N_ESTIMATORS
number of trees in the forest
-f N_FEATURES, --n_features N_FEATURES
number of features
-b BATCH_SIZE, --batch_size BATCH_SIZE
batch size
-n NUMBER, --number NUMBER
number of calls to measure
-w WARMUP, --warmup WARMUP
warmup calls before starting to measure
-r REPEAT, --repeat REPEAT
number of measures to repeat
-e ENGINE, --engine ENGINE
implementation to checks, comma separated list,
possible values onnxruntime,onnxruntime-
customops,CReferenceEvaluator,cython,cython-customops
-v, --verbose verbose, default is False
-p, --profile run a profiling
-o OUTPUT, --output OUTPUT
output file to write
See function onnx_extended.validation.bench_trees.bench_trees().
check#
The function checks the package is working after it was installed. It shortly run a few functions to check shared libraries can be used by the package.
usage: check [-h] [-o] [-a] [-r] [-v]
Quickly checks the module is properly installed.
options:
-h, --help show this help message and exit
-o, --ortcy check OrtSession
-a, --val check submodule validation
-r, --ortops check custom operators with onnxruntime
-v, --verbose verbose, default is False
cvt#
Conversion of a file into another format, usually a csv file into an excel file.
usage: stat [-h] -i INPUT -o OUTPUT [-v]
Converts a file into another format, usually a csv file into an excel file.
The extension defines the conversion.
options:
-h, --help show this help message and exit
-i INPUT, --input INPUT
input file
-o OUTPUT, --output OUTPUT
output file
-v, --verbose verbose, default is False
display#
Displays information from the shape inference on the standard output and in a csv file.
usage: display [-h] -m MODEL [-s SAVE] [-t TAB] [-e EXTERNAL]
Executes shape inference on an ONNX model and displays the inferred shapes.
options:
-h, --help show this help message and exit
-m MODEL, --model MODEL
onnx model to display
-s SAVE, --save SAVE saved the data as a dataframe
-t TAB, --tab TAB column size when printed on standard output
-e EXTERNAL, --external EXTERNAL
load external data?
This helps looking at a model from a terminal.
Output example:
input tensor X FLOAT ?x?
input tensor Y FLOAT 5x6
Op Add , FLOAT X,Y res
result tensor res FLOAT 5x6
Op Cos FLOAT FLOAT res Z
result tensor Z FLOAT ?x?
output tensor Z FLOAT ?x?
- onnx_extended._command_lines.display_intermediate_results(model: str, save: str | None = None, tab: int = 12, external: bool = True, fprint: ~typing.Callable = <built-in function print>)[source]#
Displays shape, type for a model.
- Parameters:
model – a model
save – save the results as a dataframe
tab – column size for the output
external – loads the external data or not
fprint – function to print
external#
Split the model and the coefficients. The coefficients goes to an external file.
usage: external [-h] -m MODEL -s SAVE [-v]
Takes an onnx model and split the model and the coefficients.
options:
-h, --help show this help message and exit
-m MODEL, --model MODEL
onnx model
-s SAVE, --save SAVE saves into that file
-v, --verbose display sizes
The functions stores the coefficients as external data. It calls the function
convert_model_to_external_data.
plot#
Plots a graph like a profiling.
usage: plot [-h] -k {profile_op,profile_node} -i INPUT [-c OCSV] [-o OPNG]
[-v] [-w] [-t TITLE]
Plots a graph representing the data loaded from a profiling stored in a
filename.
options:
-h, --help show this help message and exit
-k {profile_op,profile_node}, --kind {profile_op,profile_node}
Kind of plot to draw. 'profile_op' shows the time
spent in every kernel per operator type,
'profile_node' shows the time spent in every kernel
per operator node,
-i INPUT, --input INPUT
input file
-c OCSV, --ocsv OCSV saves the data used to plot the graph as csv file
-o OPNG, --opng OPNG saves the plot as png into that file
-v, --verbose display sizes
-w, --with-shape keep the shape before aggregating the results
-t TITLE, --title TITLE
plot title
Plots a graph
Output example:
[plot_profile] load 'onnxruntime_profile__2023-11-02_10-07-44.json'
args_node_index,args_op_name,args_provider,it==0,dur,ratio
1,Cos,CPU,1,11,0.08396946564885496
1,Cos,CPU,0,29,0.22137404580152673
0,Neg,CPU,1,46,0.3511450381679389
0,Neg,CPU,0,45,0.3435114503816794
[plot_profile] save '/tmp/tmp40_jc95t/o.csv'
[plot_profile] save '/tmp/tmp40_jc95t/o.png'
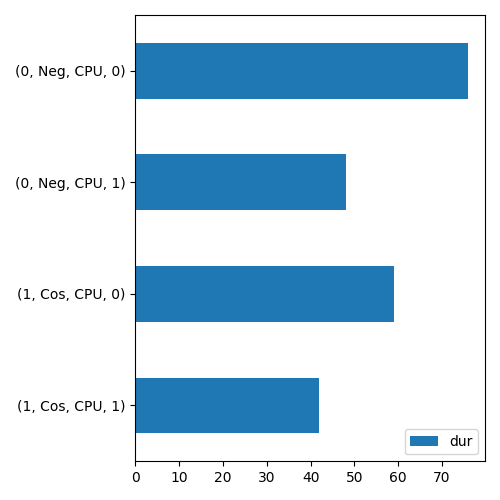
- onnx_extended._command_lines.cmd_plot(filename: str, kind: str, out_csv: str | None = None, out_png: str | None = None, title: str | None = None, with_shape: bool = False, verbose: int = 0)[source]#
Plots a graph.
- Parameters:
filename – raw data to load
kind – kind of plot to so, see below
out_csv – output the data into that csv file
out_png – output the graph in that file
title – title (optional)
with_shape – keep the shape to aggregate
verbose – verbosity, if > 0, prints out the data in csv format
Kinds of plots:
‘profile_op’: draws the profiling per node type
‘profile_node’: draws the profiling per node
print#
Prints a model or a tensor on the standard output.
usage: print [-h] -i INPUT [-f {raw,nodes,opsets}] [-e EXTERNAL]
Shows an onnx model or a protobuf string on stdout. Extension '.onnx' is
considered a model, extension '.proto' or '.pb' is a protobuf string.
options:
-h, --help show this help message and exit
-i INPUT, --input INPUT
onnx model or protobuf file to print
-f {raw,nodes,opsets}, --format {raw,nodes,opsets}
format to use to display the graph, 'raw' means the
json-like format, 'nodes' shows all the nodes, input
and outputs in the main graph, 'opsets' shows the
opsets and ir_version
-e EXTERNAL, --external EXTERNAL
load external data?
The command can be used on short models, mostly coming from unittests. Big
models are far too large to make this command useful. Use command display
instead.
Output example:
Type: <class 'onnx.onnx_ml_pb2.ModelProto'>
ir_version: 9
opset_import {
domain: ""
version: 18
}
graph {
node {
input: "X"
input: "Y"
output: "res"
op_type: "Add"
}
node {
input: "res"
output: "Z"
op_type: "Cos"
}
name: "g"
input {
name: "X"
type {
tensor_type {
elem_type: 1
shape {
dim {
}
dim {
}
}
}
}
}
input {
name: "Y"
type {
tensor_type {
elem_type: 1
shape {
dim {
dim_value: 5
}
dim {
dim_value: 6
}
}
}
}
}
output {
name: "Z"
type {
tensor_type {
elem_type: 1
shape {
dim {
}
dim {
}
}
}
}
}
}
- onnx_extended._command_lines.print_proto(proto: str, fmt: str = 'raw', external: bool = True)[source]#
Shows an onnx model or a protobuf string on stdout. Extension ‘.onnx’ is considered a model, extension ‘.proto’ or ‘.pb’ is a protobuf string.
- Parameters:
proto – a file
fmt – format to use to print the model, raw prints out the string produced by print(model), nodes only prints out the node name
external – loads with external data
quantize#
Prints a model or a tensor on the standard output.
usage: quantize [-h] -i INPUT -o OUTPUT -k {fp8,fp16}
[-s {onnxruntime,onnx-extended}] [-e EARLY_STOP] [-q] [-v]
[-t TRANSPOSE] [-x EXCLUDE] [-p OPTIONS]
Quantizes a model in simple ways.
options:
-h, --help show this help message and exit
-i INPUT, --input INPUT
onnx model or protobuf file to print
-o OUTPUT, --output OUTPUT
output model to write
-k {fp8,fp16}, --kind {fp8,fp16}
Kind of quantization to do. 'fp8' quantizes weights to
float 8 e4m3fn whenever possible. It replaces MatMul
by Transpose + DynamicQuantizeLinear + GemmFloat8.
'fp16' casts all float weights to float 16, it does
the same for inputs and outputs. It changes all
operators Cast when they cast into float 32.
-s {onnxruntime,onnx-extended}, --scenario {onnxruntime,onnx-extended}
Possible versions for fp8 quantization. 'onnxruntime'
uses operators implemented by onnxruntime, 'onnx-
extended' uses experimental operators from this
package.
-e EARLY_STOP, --early-stop EARLY_STOP
stops after N modifications
-q, --quiet do not stop if an exception is raised
-v, --verbose enable logging, can be repeated
-t TRANSPOSE, --transpose TRANSPOSE
which input(s) to transpose, 1 for the first one, 2
for the second, 3 for both, 0 for None
-x EXCLUDE, --exclude EXCLUDE
to avoid quantizing nodes if their names belongs to
that list (comma separated)
-p OPTIONS, --options OPTIONS
options to use for quantization, NONE (default) or
OPTIMIZE, several values can be passed separated by a
comma
The implementation quantization are mostly experimental. Once finalized, the
functionality might move to another package.
Example:
python3 -m onnx_extended quantize -i bertsquad-12.onnx -o bertsquad-12-fp8-1.onnx -v -v -k fp8 -q
Output example:
INFO:onnx-extended:Model initial size: 143
INFO:onnx-extended/transformer:[quantize_float8] upgrade model from opset 18 to 19
INFO:onnx-extended/transformer:[quantize_float8] 2/4 quantize Node(2, <parent>, <MatMul>) [X,mat] -> [Z]
INFO:onnx-extended:Model quantized size: 991
- onnx_extended._command_lines.cmd_quantize(model: ModelProto | str, output: str | None = None, kind: str = 'fp8', scenario: str = 'onnxruntime', early_stop: int | None = None, quiet: bool = False, verbose: int = 0, index_transpose: int = 2, exceptions: List[Dict[str, str]] | None = None, options: QuantizeOptions | None = None)[source]#
Quantizes a model
- Parameters:
model – path to a model or ModelProto
output – output file
kind – kind of quantization
scenario – depends on the quantization
early_stop – stops early to see the preliminary results
quiet – do not stop an exception
verbose – verbosity level
index_transpose – which input to transpose before calling gemm: 0 (none), 1 (first), 2 (second), 3 for both
exceptions – exclude nodes from the quantization, [{“name”: “node_name1”}, {“name”: “node_name2”}] will exclude these two node names from the quantization
options – quantization options, see class
QuantizeOptions
select#
Extracts a subpart of an existing model.
usage: select [-h] -m MODEL -s SAVE [-i INPUTS] [-o OUTPUTS] [-v]
[-t TRANSPOSE]
Selects a subpart of an onnx model.
options:
-h, --help show this help message and exit
-m MODEL, --model MODEL
onnx model
-s SAVE, --save SAVE saves into that file
-i INPUTS, --inputs INPUTS
list of inputs to keep, comma separated values, leave
empty to keep the model inputs
-o OUTPUTS, --outputs OUTPUTS
list of outputs to keep, comma separated values, leave
empty to keep the model outputs
-v, --verbose verbose, default is False
-t TRANSPOSE, --transpose TRANSPOSE
which input to transpose before calling gemm: 0
(none), 1 (first), 2 (second), 3 for both
The function removes the unused nodes.
Output example:
INFO:onnx-extended:Initial model size: 101
INFO:onnx-extended:[select_model_inputs_outputs] nodes 2 --> 1
INFO:onnx-extended:[select_model_inputs_outputs] inputs: ['X', 'Y']
INFO:onnx-extended:[select_model_inputs_outputs] inputs: ['res']
INFO:onnx-extended:Selected model size: 102
- onnx_extended._command_lines.cmd_select(model: ModelProto | str, save: str | None = None, inputs: str | List[str] | None = None, outputs: str | List[str] | None = None, verbose: int = 0)[source]#
Selects a subgraph in a model.
- Parameters:
model – path to a model or ModelProto
save – model ot save in this file
inputs – list of inputs or empty to keep the original inputs
outputs – list of outputs or empty to keep the original outputs
verbose – verbosity level
stat#
Produces statistics on initiliazers and tree ensemble in an onnx model.
See onnx_extended.tools.stats_nodes.enumerate_stats_nodes()
usage: stat [-h] -i INPUT -o OUTPUT [-v]
Computes statistics on initiliazer and tree ensemble nodes.
options:
-h, --help show this help message and exit
-i INPUT, --input INPUT
onnx model file name
-o OUTPUT, --output OUTPUT
file name, contains a csv file
-v, --verbose verbose, default is False
Output example:
[cmd_stat] load model '/tmp/tmpxqeqawk1/m.onnx'
[cmd_stat] object 0: name=('add', 'Y') size=4 dtype=float32
[cmd_stat] prints out /tmp/tmpxqeqawk1/stat.scsv
index joined_name size shape dtype min max mean ... hist_x_13 hist_x_14 hist_x_15 hist_x_16 hist_x_17 hist_x_18 hist_x_19 hist_x_20
0 0 add|Y 4 (2, 2) float32 2.0 5.0 3.5 ... 3.95 4.1 4.25 4.4 4.55 4.7 4.85 5.0
[1 rows x 51 columns]
Output example with trees:
index joined_name kind n_trees ... n_rules rules hist_rules__BRANCH_LEQ hist_rules__LEAF
0 0 ONNX(RandomForestRegressor)|TreeEnsembleRegressor Regressor 3 ... 2 BRANCH_LEQ,LEAF 9 12
[1 rows x 11 columns]
store#
Stores intermediate outputs on disk.
See also CReferenceEvaluator.
usage: store [-h] -m MODEL [-o OUT] -i INPUT [-v] [-r {CReferenceEvaluator}]
[-p PROVIDERS]
Executes a model with class CReferenceEvaluator and stores every intermediate
results on disk with a short onnx to execute the node.
options:
-h, --help show this help message and exit
-m MODEL, --model MODEL
onnx model to test
-o OUT, --out OUT path where to store the outputs, default is '.'
-i INPUT, --input INPUT
input, it can be a path, or a string like
'float32(4,5)' to generate a random input of this
shape, 'rnd' works as well if the model precisely
defines the inputs, 'float32(4,5):U10' generates a
tensor with a uniform law
-v, --verbose verbose, default is False
-r {CReferenceEvaluator}, --runtime {CReferenceEvaluator}
Runtime to use to generate the intermediate results,
default is 'CReferenceEvaluator'
-p PROVIDERS, --providers PROVIDERS
Execution providers, multiple values can separated
with a comma
This is inspired from PR https://github.com/onnx/onnx/pull/5413. This command
may disappear if this functionnality is not used.
- onnx_extended._command_lines.store_intermediate_results(model: ModelProto | str, inputs: List[str | ndarray | TensorProto], out: str = '.', runtime: type | str = 'CReferenceEvaluator', providers: str | List[str] = 'CPU', verbose: int = 0)[source]#
Executes an onnx model with a runtime and stores the intermediate results in a folder. See
CReferenceEvaluatorfor further details.- Parameters:
model – path to a model or ModelProto
inputs – list of inputs for the model
out – output path
runtime – runtime class to use
providers – list of providers
verbose – verbosity level
- Returns:
outputs