Note
Go to the end to download the full example code
TreeEnsemble optimization#
The execution of a TreeEnsembleRegressor can lead to very different results depending on how the computation is parallelized. By trees, by rows, by both, for only one row, for a short batch of rows, a longer one. The implementation in onnxruntime does not let the user changed the predetermined settings but a custom kernel might. That’s what this example is measuring.
The default set of optimized parameters is very short and is meant to be executed fast. Many more parameters can be tried.
python plot_op_tree_ensemble_optim --scenario=LONG
To change the training parameters:
python plot_op_tree_ensemble_optim.py
--n_trees=100
--max_depth=10
--n_features=50
--batch_size=100000
Another example with a full list of parameters:
- python plot_op_tree_ensemble_optim.py
–n_trees=100 –max_depth=10 –n_features=50 –batch_size=100000 –tries=3 –scenario=CUSTOM –parallel_tree=80,40 –parallel_tree_N=128,64 –parallel_N=50,25 –batch_size_tree=1,2 –batch_size_rows=1,2 –use_node3=0
Another example:
python plot_op_tree_ensemble_optim.py
--n_trees=100 --n_features=10 --batch_size=10000 --max_depth=8 -s SHORT
import logging
import os
import timeit
from typing import Tuple
import numpy
import onnx
from onnx import ModelProto
from onnx.helper import make_graph, make_model
from onnx.reference import ReferenceEvaluator
from pandas import DataFrame
from sklearn.datasets import make_regression
from sklearn.ensemble import RandomForestRegressor
from skl2onnx import to_onnx
from onnxruntime import InferenceSession, SessionOptions
from onnx_array_api.plotting.text_plot import onnx_simple_text_plot
from onnx_extended.reference import CReferenceEvaluator
from onnx_extended.ortops.optim.cpu import get_ort_ext_libs
from onnx_extended.ortops.optim.optimize import (
change_onnx_operator_domain,
get_node_attribute,
optimize_model,
)
from onnx_extended.tools.onnx_nodes import multiply_tree
from onnx_extended.args import get_parsed_args
from onnx_extended.ext_test_case import unit_test_going
from onnx_extended.plotting.benchmark import hhistograms
logging.getLogger("matplotlib.font_manager").setLevel(logging.ERROR)
script_args = get_parsed_args(
"plot_op_tree_ensemble_optim",
description=__doc__,
scenarios={
"SHORT": "short optimization (default)",
"LONG": "test more options",
"CUSTOM": "use values specified by the command line",
},
n_features=(2 if unit_test_going() else 5, "number of features to generate"),
n_trees=(3 if unit_test_going() else 10, "number of trees to train"),
max_depth=(2 if unit_test_going() else 5, "max_depth"),
batch_size=(1000 if unit_test_going() else 10000, "batch size"),
parallel_tree=("80,160,40", "values to try for parallel_tree"),
parallel_tree_N=("256,128,64", "values to try for parallel_tree_N"),
parallel_N=("100,50,25", "values to try for parallel_N"),
batch_size_tree=("2,4,8", "values to try for batch_size_tree"),
batch_size_rows=("2,4,8", "values to try for batch_size_rows"),
use_node3=("0,1", "values to try for use_node3"),
expose="",
n_jobs=("-1", "number of jobs to train the RandomForestRegressor"),
)
Training a model#
def train_model(
batch_size: int, n_features: int, n_trees: int, max_depth: int
) -> Tuple[str, numpy.ndarray, numpy.ndarray]:
filename = f"plot_op_tree_ensemble_optim-f{n_features}-{n_trees}-d{max_depth}.onnx"
if not os.path.exists(filename):
X, y = make_regression(
batch_size + max(batch_size, 2 ** (max_depth + 1)),
n_features=n_features,
n_targets=1,
)
print(f"Training to get {filename!r} with X.shape={X.shape}")
X, y = X.astype(numpy.float32), y.astype(numpy.float32)
# To be faster, we train only 1 tree.
model = RandomForestRegressor(
1, max_depth=max_depth, verbose=2, n_jobs=int(script_args.n_jobs)
)
model.fit(X[:-batch_size], y[:-batch_size])
onx = to_onnx(model, X[:1])
# And wd multiply the trees.
node = multiply_tree(onx.graph.node[0], n_trees)
onx = make_model(
make_graph([node], onx.graph.name, onx.graph.input, onx.graph.output),
domain=onx.domain,
opset_imports=onx.opset_import,
)
with open(filename, "wb") as f:
f.write(onx.SerializeToString())
else:
X, y = make_regression(batch_size, n_features=n_features, n_targets=1)
X, y = X.astype(numpy.float32), y.astype(numpy.float32)
Xb, yb = X[-batch_size:].copy(), y[-batch_size:].copy()
return filename, Xb, yb
batch_size = script_args.batch_size
n_features = script_args.n_features
n_trees = script_args.n_trees
max_depth = script_args.max_depth
print(f"batch_size={batch_size}")
print(f"n_features={n_features}")
print(f"n_trees={n_trees}")
print(f"max_depth={max_depth}")
batch_size=10000
n_features=5
n_trees=10
max_depth=5
training
filename, Xb, yb = train_model(batch_size, n_features, n_trees, max_depth)
print(f"Xb.shape={Xb.shape}")
print(f"yb.shape={yb.shape}")
Training to get 'plot_op_tree_ensemble_optim-f5-10-d5.onnx' with X.shape=(20000, 5)
[Parallel(n_jobs=-1)]: Using backend ThreadingBackend with 8 concurrent workers.
building tree 1 of 1
[Parallel(n_jobs=-1)]: Done 1 out of 1 | elapsed: 0.2s finished
Xb.shape=(10000, 5)
yb.shape=(10000,)
Rewrite the onnx file to use a different kernel#
The custom kernel is mapped to a custom operator with the same name the attributes and domain = “onnx_extented.ortops.optim.cpu”. We call a function to do that replacement. First the current model.
opset: domain='ai.onnx.ml' version=1
opset: domain='' version=19
input: name='X' type=dtype('float32') shape=['', 5]
TreeEnsembleRegressor(X, n_targets=1, nodes_falsenodeids=630:[32,17,10...62,0,0], nodes_featureids=630:[0,0,3...2,0,0], nodes_hitrates=630:[1.0,1.0...1.0,1.0], nodes_missing_value_tracks_true=630:[0,0,0...0,0,0], nodes_modes=630:[b'BRANCH_LEQ',b'BRANCH_LEQ'...b'LEAF',b'LEAF'], nodes_nodeids=630:[0,1,2...60,61,62], nodes_treeids=630:[0,0,0...9,9,9], nodes_truenodeids=630:[1,2,3...61,0,0], nodes_values=630:[-0.20009872317314148,-1.1906558275222778...0.0,0.0], post_transform=b'NONE', target_ids=320:[0,0,0...0,0,0], target_nodeids=320:[5,6,8...59,61,62], target_treeids=320:[0,0,0...9,9,9], target_weights=320:[-162.12649536132812,-114.27810668945312...107.13147735595703,152.6385955810547]) -> variable
output: name='variable' type=dtype('float32') shape=['', 1]
And then the modified model.
def transform_model(model, **kwargs):
onx = ModelProto()
onx.ParseFromString(model.SerializeToString())
att = get_node_attribute(onx.graph.node[0], "nodes_modes")
modes = ",".join(map(lambda s: s.decode("ascii"), att.strings)).replace(
"BRANCH_", ""
)
return change_onnx_operator_domain(
onx,
op_type="TreeEnsembleRegressor",
op_domain="ai.onnx.ml",
new_op_domain="onnx_extented.ortops.optim.cpu",
nodes_modes=modes,
**kwargs,
)
print("Tranform model to add a custom node.")
onx_modified = transform_model(onx)
print(f"Save into {filename + 'modified.onnx'!r}.")
with open(filename + "modified.onnx", "wb") as f:
f.write(onx_modified.SerializeToString())
print("done.")
print(onnx_simple_text_plot(onx_modified))
Tranform model to add a custom node.
Save into 'plot_op_tree_ensemble_optim-f5-10-d5.onnxmodified.onnx'.
done.
opset: domain='ai.onnx.ml' version=1
opset: domain='' version=19
opset: domain='onnx_extented.ortops.optim.cpu' version=1
input: name='X' type=dtype('float32') shape=['', 5]
TreeEnsembleRegressor[onnx_extented.ortops.optim.cpu](X, nodes_modes=b'LEQ,LEQ,LEQ,LEQ,LEQ,LEAF,LEAF,LEQ,LEAF...LEAF,LEAF', n_targets=1, nodes_falsenodeids=630:[32,17,10...62,0,0], nodes_featureids=630:[0,0,3...2,0,0], nodes_hitrates=630:[1.0,1.0...1.0,1.0], nodes_missing_value_tracks_true=630:[0,0,0...0,0,0], nodes_nodeids=630:[0,1,2...60,61,62], nodes_treeids=630:[0,0,0...9,9,9], nodes_truenodeids=630:[1,2,3...61,0,0], nodes_values=630:[-0.20009872317314148,-1.1906558275222778...0.0,0.0], post_transform=b'NONE', target_ids=320:[0,0,0...0,0,0], target_nodeids=320:[5,6,8...59,61,62], target_treeids=320:[0,0,0...9,9,9], target_weights=320:[-162.12649536132812,-114.27810668945312...107.13147735595703,152.6385955810547]) -> variable
output: name='variable' type=dtype('float32') shape=['', 1]
Comparing onnxruntime and the custom kernel#
print(f"Loading {filename!r}")
sess_ort = InferenceSession(filename, providers=["CPUExecutionProvider"])
r = get_ort_ext_libs()
print(f"Creating SessionOptions with {r!r}")
opts = SessionOptions()
if r is not None:
opts.register_custom_ops_library(r[0])
print(f"Loading modified {filename!r}")
sess_cus = InferenceSession(
onx_modified.SerializeToString(), opts, providers=["CPUExecutionProvider"]
)
print(f"Running once with shape {Xb.shape}.")
base = sess_ort.run(None, {"X": Xb})[0]
print(f"Running modified with shape {Xb.shape}.")
got = sess_cus.run(None, {"X": Xb})[0]
print("done.")
Loading 'plot_op_tree_ensemble_optim-f5-10-d5.onnx'
Creating SessionOptions with ['~/github/onnx-extended/onnx_extended/ortops/optim/cpu/libortops_optim_cpu.so']
Loading modified 'plot_op_tree_ensemble_optim-f5-10-d5.onnx'
Running once with shape (10000, 5).
Running modified with shape (10000, 5).
done.
Discrepancies?
Discrepancies: max=2.56771443218895e-07, mean=5.363955679626997e-08 (A=950.80908203125)
Simple verification#
Baseline with onnxruntime.
t1 = timeit.timeit(lambda: sess_ort.run(None, {"X": Xb}), number=50)
print(f"baseline: {t1}")
baseline: 0.10309480000023541
The custom implementation.
t2 = timeit.timeit(lambda: sess_cus.run(None, {"X": Xb}), number=50)
print(f"new time: {t2}")
new time: 0.05421680000017659
The same implementation but ran from the onnx python backend.
ref = CReferenceEvaluator(filename)
ref.run(None, {"X": Xb})
t3 = timeit.timeit(lambda: ref.run(None, {"X": Xb}), number=50)
print(f"CReferenceEvaluator: {t3}")
CReferenceEvaluator: 0.0474501999997301
The python implementation but from the onnx python backend.
ReferenceEvaluator: 4.078654500000084 (only 5 times instead of 50)
Time for comparison#
The custom kernel supports the same attributes as TreeEnsembleRegressor plus new ones to tune the parallelization. They can be seen in tree_ensemble.cc. Let’s try out many possibilities. The default values are the first ones.
if unit_test_going():
optim_params = dict(
parallel_tree=[40], # default is 80
parallel_tree_N=[128], # default is 128
parallel_N=[50, 25], # default is 50
batch_size_tree=[1], # default is 1
batch_size_rows=[1], # default is 1
use_node3=[0], # default is 0
)
elif script_args.scenario in (None, "SHORT"):
optim_params = dict(
parallel_tree=[80, 40], # default is 80
parallel_tree_N=[128, 64], # default is 128
parallel_N=[50, 25], # default is 50
batch_size_tree=[1], # default is 1
batch_size_rows=[1], # default is 1
use_node3=[0], # default is 0
)
elif script_args.scenario == "LONG":
optim_params = dict(
parallel_tree=[80, 160, 40],
parallel_tree_N=[256, 128, 64],
parallel_N=[100, 50, 25],
batch_size_tree=[1, 2, 4, 8],
batch_size_rows=[1, 2, 4, 8],
use_node3=[0, 1],
)
elif script_args.scenario == "CUSTOM":
optim_params = dict(
parallel_tree=list(int(i) for i in script_args.parallel_tree.split(",")),
parallel_tree_N=list(int(i) for i in script_args.parallel_tree_N.split(",")),
parallel_N=list(int(i) for i in script_args.parallel_N.split(",")),
batch_size_tree=list(int(i) for i in script_args.batch_size_tree.split(",")),
batch_size_rows=list(int(i) for i in script_args.batch_size_rows.split(",")),
use_node3=list(int(i) for i in script_args.use_node3.split(",")),
)
else:
raise ValueError(
f"Unknown scenario {script_args.scenario!r}, use --help to get them."
)
cmds = []
for att, value in optim_params.items():
cmds.append(f"--{att}={','.join(map(str, value))}")
print("Full list of optimization parameters:")
print(" ".join(cmds))
Full list of optimization parameters:
--parallel_tree=80,40 --parallel_tree_N=128,64 --parallel_N=50,25 --batch_size_tree=1 --batch_size_rows=1 --use_node3=0
Then the optimization.
def create_session(onx):
opts = SessionOptions()
r = get_ort_ext_libs()
if r is None:
raise RuntimeError("No custom implementation available.")
opts.register_custom_ops_library(r[0])
return InferenceSession(
onx.SerializeToString(), opts, providers=["CPUExecutionProvider"]
)
res = optimize_model(
onx,
feeds={"X": Xb},
transform=transform_model,
session=create_session,
baseline=lambda onx: InferenceSession(
onx.SerializeToString(), providers=["CPUExecutionProvider"]
),
params=optim_params,
verbose=True,
number=script_args.number,
repeat=script_args.repeat,
warmup=script_args.warmup,
sleep=script_args.sleep,
n_tries=script_args.tries,
)
0%| | 0/16 [00:00<?, ?it/s]
i=1/16 TRY=0 //tree=80 //tree_N=128 //N=50 bs_tree=1 batch_size_rows=1 n3=0: 0%| | 0/16 [00:00<?, ?it/s]
i=1/16 TRY=0 //tree=80 //tree_N=128 //N=50 bs_tree=1 batch_size_rows=1 n3=0: 6%|▋ | 1/16 [00:00<00:07, 1.96it/s]
i=2/16 TRY=0 //tree=80 //tree_N=128 //N=25 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 6%|▋ | 1/16 [00:00<00:07, 1.96it/s]
i=2/16 TRY=0 //tree=80 //tree_N=128 //N=25 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 12%|█▎ | 2/16 [00:00<00:05, 2.71it/s]
i=3/16 TRY=0 //tree=80 //tree_N=64 //N=50 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 12%|█▎ | 2/16 [00:00<00:05, 2.71it/s]
i=3/16 TRY=0 //tree=80 //tree_N=64 //N=50 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 19%|█▉ | 3/16 [00:01<00:04, 3.05it/s]
i=4/16 TRY=0 //tree=80 //tree_N=64 //N=25 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 19%|█▉ | 3/16 [00:01<00:04, 3.05it/s]
i=4/16 TRY=0 //tree=80 //tree_N=64 //N=25 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 25%|██▌ | 4/16 [00:01<00:03, 3.26it/s]
i=5/16 TRY=0 //tree=40 //tree_N=128 //N=50 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 25%|██▌ | 4/16 [00:01<00:03, 3.26it/s]
i=5/16 TRY=0 //tree=40 //tree_N=128 //N=50 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 31%|███▏ | 5/16 [00:01<00:03, 3.23it/s]
i=6/16 TRY=0 //tree=40 //tree_N=128 //N=25 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 31%|███▏ | 5/16 [00:01<00:03, 3.23it/s]
i=6/16 TRY=0 //tree=40 //tree_N=128 //N=25 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 38%|███▊ | 6/16 [00:01<00:02, 3.41it/s]
i=7/16 TRY=0 //tree=40 //tree_N=64 //N=50 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 38%|███▊ | 6/16 [00:01<00:02, 3.41it/s]
i=7/16 TRY=0 //tree=40 //tree_N=64 //N=50 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 44%|████▍ | 7/16 [00:02<00:02, 3.50it/s]
i=8/16 TRY=0 //tree=40 //tree_N=64 //N=25 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 44%|████▍ | 7/16 [00:02<00:02, 3.50it/s]
i=8/16 TRY=0 //tree=40 //tree_N=64 //N=25 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 50%|█████ | 8/16 [00:02<00:02, 3.53it/s]
i=9/16 TRY=1 //tree=80 //tree_N=128 //N=50 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 50%|█████ | 8/16 [00:02<00:02, 3.53it/s]
i=9/16 TRY=1 //tree=80 //tree_N=128 //N=50 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 56%|█████▋ | 9/16 [00:02<00:01, 3.59it/s]
i=10/16 TRY=1 //tree=80 //tree_N=128 //N=25 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 56%|█████▋ | 9/16 [00:02<00:01, 3.59it/s]
i=10/16 TRY=1 //tree=80 //tree_N=128 //N=25 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 62%|██████▎ | 10/16 [00:02<00:01, 3.68it/s]
i=11/16 TRY=1 //tree=80 //tree_N=64 //N=50 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 62%|██████▎ | 10/16 [00:02<00:01, 3.68it/s]
i=11/16 TRY=1 //tree=80 //tree_N=64 //N=50 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 69%|██████▉ | 11/16 [00:03<00:01, 3.74it/s]
i=12/16 TRY=1 //tree=80 //tree_N=64 //N=25 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 69%|██████▉ | 11/16 [00:03<00:01, 3.74it/s]
i=12/16 TRY=1 //tree=80 //tree_N=64 //N=25 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 75%|███████▌ | 12/16 [00:03<00:01, 3.79it/s]
i=13/16 TRY=1 //tree=40 //tree_N=128 //N=50 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 75%|███████▌ | 12/16 [00:03<00:01, 3.79it/s]
i=13/16 TRY=1 //tree=40 //tree_N=128 //N=50 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 81%|████████▏ | 13/16 [00:03<00:00, 3.76it/s]
i=14/16 TRY=1 //tree=40 //tree_N=128 //N=25 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 81%|████████▏ | 13/16 [00:03<00:00, 3.76it/s]
i=14/16 TRY=1 //tree=40 //tree_N=128 //N=25 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 88%|████████▊ | 14/16 [00:04<00:00, 3.73it/s]
i=15/16 TRY=1 //tree=40 //tree_N=64 //N=50 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 88%|████████▊ | 14/16 [00:04<00:00, 3.73it/s]
i=15/16 TRY=1 //tree=40 //tree_N=64 //N=50 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 94%|█████████▍| 15/16 [00:04<00:00, 3.72it/s]
i=16/16 TRY=1 //tree=40 //tree_N=64 //N=25 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 94%|█████████▍| 15/16 [00:04<00:00, 3.72it/s]
i=16/16 TRY=1 //tree=40 //tree_N=64 //N=25 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 100%|██████████| 16/16 [00:04<00:00, 3.66it/s]
i=16/16 TRY=1 //tree=40 //tree_N=64 //N=25 bs_tree=1 batch_size_rows=1 n3=0 ~=1.03x: 100%|██████████| 16/16 [00:04<00:00, 3.48it/s]
And the results.
df = DataFrame(res)
df.to_csv("plot_op_tree_ensemble_optim.csv", index=False)
df.to_excel("plot_op_tree_ensemble_optim.xlsx", index=False)
print(df.columns)
print(df.head(5))
Index(['average', 'deviation', 'min_exec', 'max_exec', 'repeat', 'number',
'ttime', 'context_size', 'warmup_time', 'n_exp', 'n_exp_name',
'short_name', 'TRY', 'name', 'parallel_tree', 'parallel_tree_N',
'parallel_N', 'batch_size_tree', 'batch_size_rows', 'use_node3'],
dtype='object')
average deviation min_exec max_exec repeat number ttime context_size warmup_time ... short_name TRY name parallel_tree parallel_tree_N parallel_N batch_size_tree batch_size_rows use_node3
0 0.001382 0.000080 0.001168 0.001464 10 10 0.013825 64 0.007059 ... 0,baseline 0.0 baseline NaN NaN NaN NaN NaN NaN
1 0.001346 0.000627 0.000714 0.002295 10 10 0.013465 64 0.012119 ... 0,80,128,50,1,1,0 NaN 80,128,50,1,1,0 80.0 128.0 50.0 1.0 1.0 0.0
2 0.001477 0.000939 0.000628 0.003061 10 10 0.014766 64 0.015152 ... 0,80,128,25,1,1,0 NaN 80,128,25,1,1,0 80.0 128.0 25.0 1.0 1.0 0.0
3 0.001606 0.000868 0.000659 0.002761 10 10 0.016056 64 0.011667 ... 0,80,64,50,1,1,0 NaN 80,64,50,1,1,0 80.0 64.0 50.0 1.0 1.0 0.0
4 0.001555 0.000857 0.000624 0.002691 10 10 0.015555 64 0.014027 ... 0,80,64,25,1,1,0 NaN 80,64,25,1,1,0 80.0 64.0 25.0 1.0 1.0 0.0
[5 rows x 20 columns]
Sorting#
small_df = df.drop(
[
"min_exec",
"max_exec",
"repeat",
"number",
"context_size",
"n_exp_name",
],
axis=1,
).sort_values("average")
print(small_df.head(n=10))
average deviation ttime warmup_time n_exp short_name TRY name parallel_tree parallel_tree_N parallel_N batch_size_tree batch_size_rows use_node3
1 0.001346 0.000627 0.013465 0.012119 0 0,80,128,50,1,1,0 NaN 80,128,50,1,1,0 80.0 128.0 50.0 1.0 1.0 0.0
0 0.001382 0.000080 0.013825 0.007059 0 0,baseline 0.0 baseline NaN NaN NaN NaN NaN NaN
10 0.001383 0.000805 0.013828 0.013777 9 1,80,128,25,1,1,0 NaN 80,128,25,1,1,0 80.0 128.0 25.0 1.0 1.0 0.0
11 0.001410 0.000598 0.014102 0.012231 10 1,80,64,50,1,1,0 NaN 80,64,50,1,1,0 80.0 64.0 50.0 1.0 1.0 0.0
6 0.001430 0.000618 0.014300 0.010566 5 0,40,128,25,1,1,0 NaN 40,128,25,1,1,0 40.0 128.0 25.0 1.0 1.0 0.0
12 0.001446 0.000577 0.014460 0.004873 11 1,80,64,25,1,1,0 NaN 80,64,25,1,1,0 80.0 64.0 25.0 1.0 1.0 0.0
2 0.001477 0.000939 0.014766 0.015152 1 0,80,128,25,1,1,0 NaN 80,128,25,1,1,0 80.0 128.0 25.0 1.0 1.0 0.0
9 0.001512 0.000731 0.015122 0.009226 8 1,80,128,50,1,1,0 NaN 80,128,50,1,1,0 80.0 128.0 50.0 1.0 1.0 0.0
13 0.001528 0.000717 0.015284 0.011832 12 1,40,128,50,1,1,0 NaN 40,128,50,1,1,0 40.0 128.0 50.0 1.0 1.0 0.0
7 0.001552 0.000674 0.015521 0.010729 6 0,40,64,50,1,1,0 NaN 40,64,50,1,1,0 40.0 64.0 50.0 1.0 1.0 0.0
Worst#
print(small_df.tail(n=10))
average deviation ttime warmup_time n_exp short_name TRY name parallel_tree parallel_tree_N parallel_N batch_size_tree batch_size_rows use_node3
13 0.001528 0.000717 0.015284 0.011832 12 1,40,128,50,1,1,0 NaN 40,128,50,1,1,0 40.0 128.0 50.0 1.0 1.0 0.0
7 0.001552 0.000674 0.015521 0.010729 6 0,40,64,50,1,1,0 NaN 40,64,50,1,1,0 40.0 64.0 50.0 1.0 1.0 0.0
4 0.001555 0.000857 0.015555 0.014027 3 0,80,64,25,1,1,0 NaN 80,64,25,1,1,0 80.0 64.0 25.0 1.0 1.0 0.0
14 0.001570 0.000872 0.015696 0.011788 13 1,40,128,25,1,1,0 NaN 40,128,25,1,1,0 40.0 128.0 25.0 1.0 1.0 0.0
8 0.001583 0.000816 0.015826 0.014267 7 0,40,64,25,1,1,0 NaN 40,64,25,1,1,0 40.0 64.0 25.0 1.0 1.0 0.0
3 0.001606 0.000868 0.016056 0.011667 2 0,80,64,50,1,1,0 NaN 80,64,50,1,1,0 80.0 64.0 50.0 1.0 1.0 0.0
15 0.001616 0.000736 0.016158 0.005087 14 1,40,64,50,1,1,0 NaN 40,64,50,1,1,0 40.0 64.0 50.0 1.0 1.0 0.0
16 0.001713 0.000969 0.017125 0.007210 15 1,40,64,25,1,1,0 NaN 40,64,25,1,1,0 40.0 64.0 25.0 1.0 1.0 0.0
17 0.001937 0.001396 0.019371 0.022326 0 1,baseline 1.0 baseline NaN NaN NaN NaN NaN NaN
5 0.001968 0.000770 0.019678 0.012655 4 0,40,128,50,1,1,0 NaN 40,128,50,1,1,0 40.0 128.0 50.0 1.0 1.0 0.0
Plot#
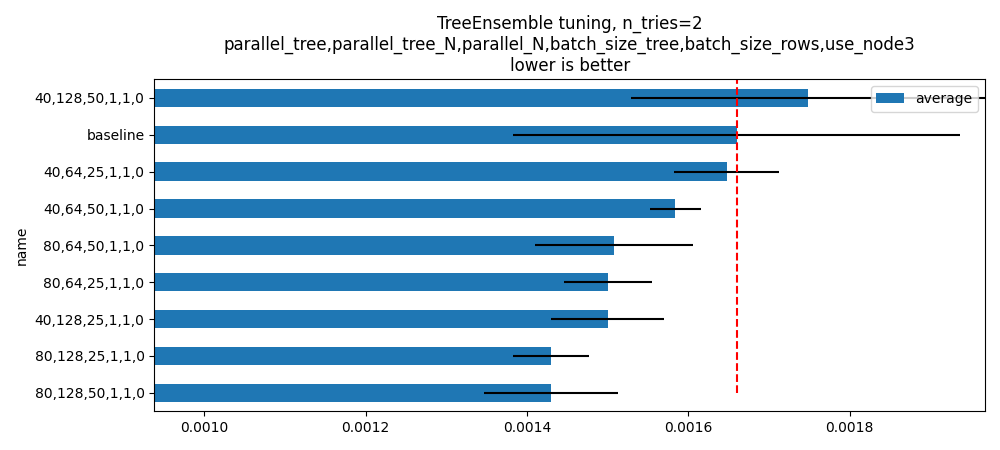
skeys = ",".join(optim_params.keys())
title = f"TreeEnsemble tuning, n_tries={script_args.tries}\n{skeys}\nlower is better"
ax = hhistograms(df, title=title, keys=("name",))
fig = ax.get_figure()
fig.savefig("plot_op_tree_ensemble_optim.png")
Total running time of the script: (0 minutes 10.922 seconds)