Note
Go to the end to download the full example code
Evaluate different implementation of TreeEnsemble#
This is a simplified benchmark to compare TreeEnsemble implementations (see below) Run python plot_op_tree_ensemble_implementations.py –help to change the tree dimension. Here are the following implementation:
ort: current onnxruntime implementations
custom: very close implementation of TreeEnsemble from onnxruntime, it allows more options to parallelize. The default is to use the parallelization settings as onnxruntime.
cusopt: it calls the same implementations as custom but with parallelization settings defined through the command line. These settings can be optimized with function
onnx_extended.ortops.optim.optimize.optimize_model(). It is usually possible to gain 10% to 20%.sparse: the input matrix used for this test can be as sparse as desired. The custom implementations can leverage this sparsity. It reduces the memory peak but it is usually slower and a dense representation of the features.
assembly: the tree is compiled with TreeBeard and this assembly is called though a custom kernel implemented for this only purpose. The tree is compiled for a particular machine and once it is compiled, the batch size cannot be changed any more. That’s why this benchmark only compares one configuration specified in the command line arguments.
Sparse Data#
import logging
import pickle
import os
import subprocess
import multiprocessing
from typing import Any, Dict, Iterator, Optional, Tuple, Union
import warnings
import numpy
import matplotlib.pyplot as plt
import onnx
from onnx import ModelProto, TensorProto
from onnx.helper import make_attribute, make_graph, make_model, make_tensor_value_info
from onnx.reference import ReferenceEvaluator
from pandas import DataFrame
from sklearn.datasets import make_regression
from sklearn.ensemble import RandomForestRegressor
from skl2onnx import to_onnx
from onnxruntime import InferenceSession, SessionOptions
from onnx_extended.ortops.optim.cpu import get_ort_ext_libs
from onnx_extended.ortops.optim.optimize import (
change_onnx_operator_domain,
get_node_attribute,
)
from onnx_extended.tools.onnx_nodes import multiply_tree
from onnx_extended.validation.cpu._validation import dense_to_sparse_struct
# from onnx_extended.plotting.benchmark import hhistograms
from onnx_extended.args import get_parsed_args
from onnx_extended.ext_test_case import unit_test_going
from onnx_extended.ext_test_case import measure_time
logging.getLogger("matplotlib.font_manager").setLevel(logging.ERROR)
script_args = get_parsed_args(
"plot_op_tree_ensemble_sparse",
description=__doc__,
scenarios={
"SHORT": "short optimization (default)",
"LONG": "test more options",
"CUSTOM": "use values specified by the command line",
},
sparsity=(0.99, "input sparsity"),
n_features=(2 if unit_test_going() else 512, "number of features to generate"),
n_trees=(3 if unit_test_going() else 512, "number of trees to train"),
max_depth=(2 if unit_test_going() else 12, "max_depth"),
batch_size=(1024 if unit_test_going() else 2048, "batch size"),
warmup=1 if unit_test_going() else 3,
parallel_tree=(128, "values to try for parallel_tree"),
parallel_tree_N=(256, "values to try for parallel_tree_N"),
parallel_N=(64, "values to try for parallel_N"),
batch_size_tree=(4, "values to try for batch_size_tree"),
batch_size_rows=(4, "values to try for batch_size_rows"),
train_all_trees=(
False,
"train all trees or replicate the first tree with a "
"random permutation of the threshold",
),
use_node3=(0, "values to try for use_node3"),
expose="",
n_jobs=("-1", "number of jobs to train the RandomForestRegressor"),
)
Training a model#
def train_model(
batch_size: int,
n_features: int,
n_trees: int,
max_depth: int,
sparsity: float,
train_all_trees: bool = False,
) -> Tuple[str, numpy.ndarray, numpy.ndarray]:
filename = (
f"plot_op_tree_ensemble_sparse-f{n_features}-{n_trees}-"
f"d{max_depth}-s{sparsity}-{1 if train_all_trees else 0}.onnx"
)
if not os.path.exists(filename):
X, y = make_regression(
batch_size + 2 ** (max_depth + 1),
n_features=n_features,
n_targets=1,
)
y -= y.mean()
y /= y.std()
mask = numpy.random.rand(*X.shape) <= sparsity
X[mask] = 0
X, y = X.astype(numpy.float32), y.astype(numpy.float32)
print(f"Training to get {filename!r} with X.shape={X.shape}")
# To be faster, we train only 1 tree.
if train_all_trees:
model = RandomForestRegressor(
n_trees, max_depth=max_depth, verbose=2, n_jobs=int(script_args.n_jobs)
)
model.fit(X[:-batch_size], y[:-batch_size])
onx = to_onnx(model, X[:1])
skl_name = filename + ".pkl"
with open(skl_name, "wb") as f:
pickle.dump(model, f)
else:
model = RandomForestRegressor(
1, max_depth=max_depth, verbose=2, n_jobs=int(script_args.n_jobs)
)
model.fit(X[:-batch_size], y[:-batch_size])
onx = to_onnx(model, X[:1])
# And wd multiply the trees.
node = multiply_tree(onx.graph.node[0], n_trees)
onx = make_model(
make_graph([node], onx.graph.name, onx.graph.input, onx.graph.output),
domain=onx.domain,
opset_imports=onx.opset_import,
)
model = None
with open(filename, "wb") as f:
f.write(onx.SerializeToString())
else:
X, y = make_regression(batch_size, n_features=n_features, n_targets=1)
mask = numpy.random.rand(*X.shape) <= sparsity
X[mask] = 0
X, y = X.astype(numpy.float32), y.astype(numpy.float32)
skl_name = filename + ".pkl"
if os.path.exists(skl_name):
with open(skl_name, "rb") as f:
model = pickle.load(f)
else:
model = None
Xb, yb = X[-batch_size:].copy(), y[-batch_size:].copy()
return filename, Xb, yb, model
def measure_sparsity(x):
f = x.flatten()
return float((f == 0).astype(numpy.int64).sum()) / float(x.size)
batch_size = script_args.batch_size
n_features = script_args.n_features
n_trees = script_args.n_trees
max_depth = script_args.max_depth
sparsity = script_args.sparsity
warmup = script_args.warmup
train_all_trees = script_args.train_all_trees in (1, "1", True, "True")
print(f"batch_size={batch_size}")
print(f"n_features={n_features}")
print(f"n_trees={n_trees}")
print(f"max_depth={max_depth}")
print(f"sparsity={sparsity}")
print(f"warmup={warmup}")
print(f"train_all_trees={train_all_trees} - {script_args.train_all_trees!r}")
filename, Xb, yb, model_skl = train_model(
batch_size,
n_features,
n_trees,
max_depth,
sparsity,
train_all_trees=train_all_trees,
)
print(f"Xb.shape={Xb.shape}")
print(f"yb.shape={yb.shape}")
print(f"measured sparsity={measure_sparsity(Xb)}")
batch_size=2048
n_features=512
n_trees=512
max_depth=12
sparsity=0.99
warmup=3
train_all_trees=False - False
Training to get 'plot_op_tree_ensemble_sparse-f512-512-d12-s0.99-0.onnx' with X.shape=(10240, 512)
[Parallel(n_jobs=-1)]: Using backend ThreadingBackend with 8 concurrent workers.
building tree 1 of 1
[Parallel(n_jobs=-1)]: Done 1 out of 1 | elapsed: 0.7s finished
Xb.shape=(2048, 512)
yb.shape=(2048,)
measured sparsity=0.9899501800537109
Implementations#
def compile_tree(
llc_exe: str,
filename: str,
onx: ModelProto,
batch_size: int,
n_features: int,
tree_tile_size: int = 8,
pipeline_width: int = 8,
reorder_tree_by_depth: bool = True,
representation_type: str = "sparse",
n_cores: Optional[int] = None,
verbose: int = 0,
) -> str:
"""
Compiles a tree with `TreeBeard <https://github.com/asprasad/treebeard>`_.
:param llc_exe: path to `llc <https://llvm.org/docs/CommandGuide/llc.html>`_
executable
:param filename: assembly name, the outcome of the compilation
:param onx: model to compile, it should contain only one node with a
TreeEssembleRegressor.
:param batch_size: batch size
:param n_features: number of features as it cannot be guessed only from the
tree definition
:param tree_tile_size: compilation parameters
:param pipeline_width: compilation parameters
:param reorder_tree_by_depth: compilation parameters
:param representation_type: compilation parameters
:param n_cores: optimized for this number of cores,
if unspecified, it uses `multiprocessing.cpu_count()`
:param verbose: to show some progress
:return: path to the generated assembly
"""
if verbose:
print("[compile_tree] import treebeard")
import treebeard
if verbose:
print(
f"[compile_tree] treebeard set options, "
f"batch_size={batch_size}, tree_tile_size={tree_tile_size}"
)
compiler_options = treebeard.CompilerOptions(batch_size, tree_tile_size)
compiler_options.SetNumberOfCores(n_cores or multiprocessing.cpu_count())
compiler_options.SetMakeAllLeavesSameDepth(pipeline_width)
compiler_options.SetReorderTreesByDepth(reorder_tree_by_depth)
compiler_options.SetNumberOfFeatures(n_features)
assert 8 < batch_size
compiler_options.SetPipelineWidth(8)
if verbose:
print(f"[compile_tree] write filename={filename!r}")
# let's remove nodes_hitrates to avoid a warning before saving the model
for node in onx.graph.node:
if node.op_type == "TreeEnsembleRegressor":
found = -1
for i in range(len(node.attribute)):
if node.attribute[i].name == "nodes_hitrates":
found = i
if found >= 0:
del node.attribute[found]
with open(filename, "wb") as f:
f.write(onx.SerializeToString())
onnx_model_path = os.path.abspath(filename)
if verbose:
print(
f"[compile_tree] treebeard context with onnx_model_path={onnx_model_path!r}"
)
tbContext = treebeard.TreebeardContext(onnx_model_path, "", compiler_options)
tbContext.SetRepresentationType(representation_type)
tbContext.SetInputFiletype("onnx_file")
llvm_file_path = f"{os.path.splitext(onnx_model_path)[0]}.ll"
if verbose:
print(f"[compile_tree] LLVM dump into {llvm_file_path!r}")
error = tbContext.DumpLLVMIR(llvm_file_path)
if error:
raise RuntimeError(
f"Failed to dump LLVM IR in {llvm_file_path!r}, error={error}."
)
if not os.path.exists(llvm_file_path):
raise FileNotFoundError(f"Unable to find {llvm_file_path!r}.")
# Run LLC
asm_file_path = f"{os.path.splitext(onnx_model_path)[0]}.s"
if verbose:
print(f"[compile_tree] llc={llc_exe!r}")
print(f"[compile_tree] run LLC into {llvm_file_path!r}")
subprocess.run(
[
llc_exe,
llvm_file_path,
"-O3",
"-march=x86-64",
"-mcpu=native",
"--relocation-model=pic",
"-o",
asm_file_path,
]
)
# Run CLANG
so_file_path = f"{os.path.splitext(onnx_model_path)[0]}.so"
if verbose:
print(f"[compile_tree] run clang into {so_file_path!r}")
subprocess.run(
["clang", "-shared", asm_file_path, "-fopenmp=libomp", "-o", so_file_path]
)
if verbose:
print("[compile_tree] done.")
return so_file_path
def make_ort_assembly_session(
onx: ModelProto, batch_size: int, n_features: int, verbose: bool = False, **kwargs
) -> Any:
"""
Creates an instance of `onnxruntime.InferenceSession` using an assembly generated
by `TreeBeard <https://github.com/asprasad/treebeard>`_.
:param onx: model to compile
:param batch_size: batch size
:param n_features: number of features as it cannot be guessed only from the
tree definition
:param verbose: verbosity
:param kwargs: any additional parameters sent to function `compile_tree`
:return: `onnxruntime.InferenceSession`
"""
from onnxruntime import InferenceSession, SessionOptions
from onnx_extended.ortops.tutorial.cpu import get_ort_ext_libs as lib_tuto
llc_exe = os.environ.get("TEST_LLC_EXE", "SKIP")
if llc_exe == "SKIP":
warnings.warn("Unable to find environment variable 'TEST_LLC_EXE'.")
return None
filename = "plot_op_tree_ensemble_implementation.onnx"
with open(filename, "wb") as f:
f.write(onx.SerializeToString())
onx = onnx.load(filename)
assembly_name = compile_tree(
llc_exe, filename, onx, batch_size, n_features, verbose=verbose, **kwargs
)
# assembly
print("change")
for node in onx.graph.node:
if node.op_type == "TreeEnsembleRegressor":
node.op_type = "TreeEnsembleAssemblyRegressor"
node.domain = "onnx_extented.ortops.tutorial.cpu"
del node.attribute[:]
new_add = make_attribute("assembly", assembly_name)
node.attribute.append(new_add)
d = onx.opset_import.add()
d.domain = "onnx_extented.ortops.tutorial.cpu"
d.version = 1
r = lib_tuto()
opts = SessionOptions()
opts.register_custom_ops_library(r[0])
sess_assembly = InferenceSession(
onx.SerializeToString(), opts, providers=["CPUExecutionProvider"]
)
return sess_assembly
def transform_model(model, use_sparse=False, **kwargs):
onx = ModelProto()
onx.ParseFromString(model.SerializeToString())
att = get_node_attribute(onx.graph.node[0], "nodes_modes")
modes = ",".join(map(lambda s: s.decode("ascii"), att.strings)).replace(
"BRANCH_", ""
)
if use_sparse and "new_op_type" not in kwargs:
kwargs["new_op_type"] = "TreeEnsembleRegressorSparse"
if use_sparse:
# with sparse tensor, missing value means 0
att = get_node_attribute(onx.graph.node[0], "nodes_values")
thresholds = numpy.array(att.floats, dtype=numpy.float32)
missing_true = (thresholds >= 0).astype(numpy.int64)
kwargs["nodes_missing_value_tracks_true"] = missing_true
new_onx = change_onnx_operator_domain(
onx,
op_type="TreeEnsembleRegressor",
op_domain="ai.onnx.ml",
new_op_domain="onnx_extented.ortops.optim.cpu",
nodes_modes=modes,
**kwargs,
)
if use_sparse:
del new_onx.graph.input[:]
new_onx.graph.input.append(
make_tensor_value_info("X", TensorProto.FLOAT, (None,))
)
return new_onx
def enumerate_implementations(
onx: ModelProto,
X: "Tensor", # noqa: F821
parallel_settings: Optional[Dict[str, int]] = None,
treebeard_settings: Optional[Dict[str, Union[int, str]]] = None,
verbose: bool = False,
) -> Iterator[Tuple[str, "onnxruntime.InferenceSession", "Tensor"]]: # noqa: F821
"""
Creates all the InferenceSession.
:param onx: model
:param X: example of an input tensor, dimension should not change
:param parallel_settings: parallelisation settings for *cusopt*, *sparse*
:param treebeard_settings: settings for treebeard compilation
:return: see annotation
"""
providers = ["CPUExecutionProvider"]
yield (
"ort",
InferenceSession(onx.SerializeToString(), providers=providers),
X,
)
r = get_ort_ext_libs()
opts = SessionOptions()
if r is not None:
opts.register_custom_ops_library(r[0])
tr = transform_model(onx)
yield (
"custom",
InferenceSession(tr.SerializeToString(), opts, providers=providers),
X,
)
tr = transform_model(onx, **parallel_settings)
yield (
"cusopt",
InferenceSession(tr.SerializeToString(), opts, providers=providers),
X,
)
Xsp = dense_to_sparse_struct(X)
tr = transform_model(onx, use_sparse=True, **parallel_settings)
yield (
"sparse",
InferenceSession(tr.SerializeToString(), opts, providers=providers),
Xsp,
)
sess = make_ort_assembly_session(
onx,
batch_size=X.shape[0],
n_features=X.shape[1],
verbose=verbose,
**treebeard_settings,
)
yield ("assembly", sess, X)
parallel_settings = dict(
parallel_tree=40,
parallel_tree_N=128,
parallel_N=50,
batch_size_tree=4,
batch_size_rows=4,
use_node3=0,
)
treebeard_settings = dict()
onx = onnx.load(filename)
sessions = []
print("----- warmup")
for name, sess, tensor in enumerate_implementations(
onx,
Xb,
parallel_settings=parallel_settings,
treebeard_settings=treebeard_settings,
verbose=1 if __name__ == "__main__" else 0,
):
if sess is None:
continue
sessions.append((name, sess, tensor))
print(f"run {name!r} - shape={tensor.shape}")
feeds = {"X": tensor}
sess.run(None, feeds)
print("done.")
----- warmup
run 'ort' - shape=(2048, 512)
run 'custom' - shape=(2048, 512)
run 'cusopt' - shape=(2048, 512)
run 'sparse' - shape=(21132,)
~/github/onnx-extended/_doc/examples/plot_op_tree_ensemble_implementations.py:345: UserWarning: Unable to find environment variable 'TEST_LLC_EXE'.
warnings.warn("Unable to find environment variable 'TEST_LLC_EXE'.")
done.
Benchmark implementations#
data = []
baseline = None
if model_skl:
print("computing the expected values with scikit-learn")
expected_values = model_skl.predict(Xb)
else:
print("computing the expected values with ReferenceEvaluator")
ref = ReferenceEvaluator(onx)
expected_values = ref.run(None, {"X": Xb})[0]
print("----- measure time")
for name, sess, tensor in sessions:
print(f"run {name!r}")
feeds = {"X": tensor}
output = sess.run(None, feeds)[0]
if baseline is None:
baseline = output
disc = 0
max_disc = 0
else:
diff = numpy.abs(output - baseline).ravel()
disc = diff.mean()
max_disc = diff.max()
obs = measure_time(
lambda: sess.run(None, feeds),
repeat=script_args.repeat,
number=script_args.number,
warmup=script_args.warmup,
)
obs["name"] = name
obs["disc_mean"] = disc
obs["disc_max"] = max_disc
diff = numpy.abs(output.ravel() - expected_values.ravel())
obs["err_mean"] = diff.mean()
obs["err_max"] = diff.max()
data.append(obs)
print("done.")
df = DataFrame(data)
print(df)
computing the expected values with ReferenceEvaluator
----- measure time
run 'ort'
run 'custom'
run 'cusopt'
run 'sparse'
done.
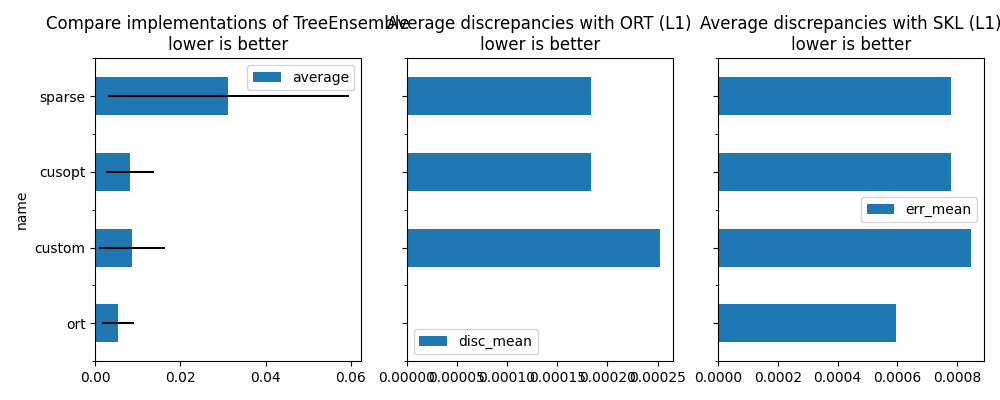
average deviation min_exec max_exec repeat number ttime context_size warmup_time name disc_mean disc_max err_mean err_max
0 0.005322 0.001205 0.003687 0.007009 10 10 0.053224 64 0.014425 ort 0.000000 0.000000 0.000596 0.000763
1 0.008599 0.000448 0.007806 0.009277 10 10 0.085989 64 0.027209 custom 0.000253 0.000488 0.000849 0.001129
2 0.008160 0.001004 0.005535 0.009274 10 10 0.081596 64 0.027325 cusopt 0.000184 0.000366 0.000780 0.001099
3 0.031239 0.001417 0.028255 0.033099 10 10 0.312390 64 0.097555 sparse 0.000184 0.000366 0.000780 0.001099
Plots.
has_skl = "err_mean" in df.columns
fig, ax = plt.subplots(1, 3 if has_skl else 2, figsize=(10, 4), sharey=True)
df[["name", "average"]].set_index("name").plot.barh(
ax=ax[0],
title="Compare implementations of TreeEnsemble\nlower is better",
xerr=[df["min_exec"], df["max_exec"]],
)
df[["name", "disc_mean"]].set_index("name").plot.barh(
ax=ax[1],
title="Average discrepancies with ORT (L1)\nlower is better",
xerr=[df["disc_max"].values * 0, df["disc_max"].values],
)
if has_skl:
df[["name", "err_mean"]].set_index("name").plot.barh(
ax=ax[2],
title="Average discrepancies with SKL (L1)\nlower is better",
xerr=[df["err_max"].values * 0, df["err_max"].values],
)
fig.tight_layout()
fig.savefig("plot_tree_ensemble_implementations.png")
Total running time of the script: (0 minutes 15.983 seconds)