Note
Go to the end to download the full example code
Measuring CPU performance with a vector sum#
The example compares the time spend in computing the sum of all coefficients of a matrix when the function walks through the coefficients by rows or by columns.
Vector Sum#
from tqdm import tqdm
import numpy
import matplotlib.pyplot as plt
from pandas import DataFrame
from onnx_extended.ext_test_case import measure_time, unit_test_going
from onnx_extended.validation.cpu._validation import vector_sum_array as vector_sum
obs = []
dims = [500, 700, 800, 900, 1000, 1100, 1200, 1300, 1400, 1500, 1600, 1700, 1800, 2000]
if unit_test_going():
dims = dims[:3]
for dim in tqdm(dims):
values = numpy.ones((dim, dim), dtype=numpy.float32).ravel()
diff = abs(vector_sum(dim, values, True) - dim**2)
res = measure_time(lambda: vector_sum(dim, values, True), max_time=0.5)
obs.append(
dict(
dim=dim,
size=values.size,
time=res["average"],
direction="rows",
time_per_element=res["average"] / dim**2,
diff=diff,
)
)
diff = abs(vector_sum(dim, values, False) - dim**2)
res = measure_time(lambda: vector_sum(dim, values, False), max_time=0.5)
obs.append(
dict(
dim=dim,
size=values.size,
time=res["average"],
direction="cols",
time_per_element=res["average"] / dim**2,
diff=diff,
)
)
df = DataFrame(obs)
piv = df.pivot(index="dim", columns="direction", values="time_per_element")
print(piv)
0%| | 0/14 [00:00<?, ?it/s]
7%|7 | 1/14 [00:01<00:16, 1.28s/it]
14%|#4 | 2/14 [00:02<00:15, 1.25s/it]
21%|##1 | 3/14 [00:03<00:14, 1.28s/it]
29%|##8 | 4/14 [00:04<00:11, 1.19s/it]
36%|###5 | 5/14 [00:06<00:11, 1.22s/it]
43%|####2 | 6/14 [00:07<00:09, 1.21s/it]
50%|##### | 7/14 [00:08<00:08, 1.16s/it]
57%|#####7 | 8/14 [00:09<00:07, 1.24s/it]
64%|######4 | 9/14 [00:11<00:06, 1.27s/it]
71%|#######1 | 10/14 [00:12<00:05, 1.30s/it]
79%|#######8 | 11/14 [00:13<00:03, 1.30s/it]
86%|########5 | 12/14 [00:14<00:02, 1.23s/it]
93%|#########2| 13/14 [00:16<00:01, 1.22s/it]
100%|##########| 14/14 [00:17<00:00, 1.23s/it]
100%|##########| 14/14 [00:17<00:00, 1.24s/it]
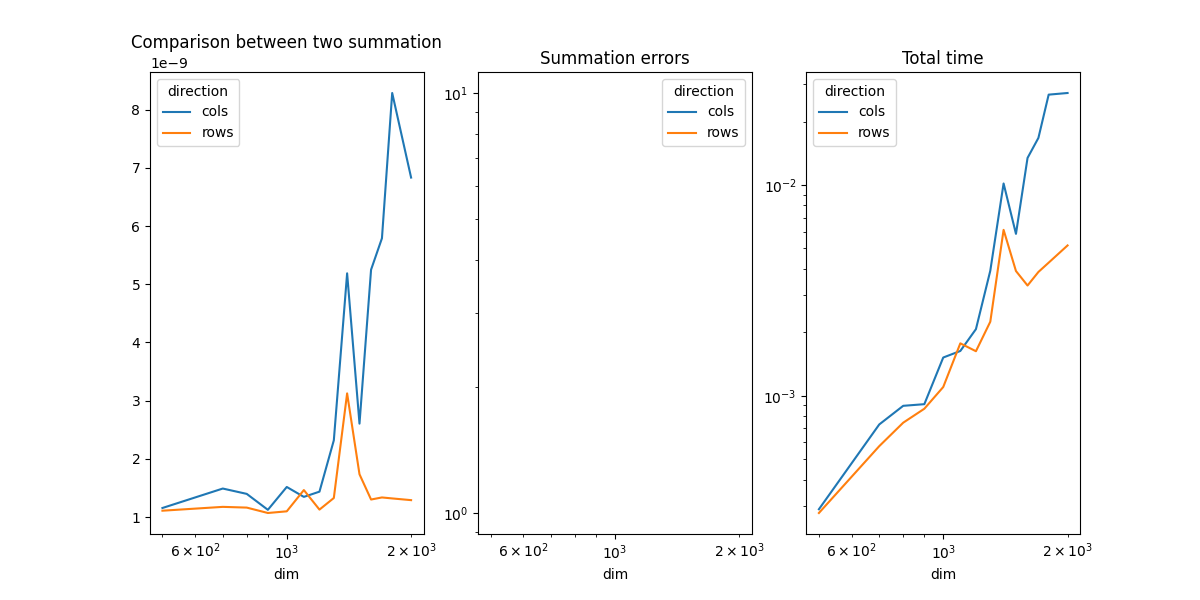
direction cols rows
dim
500 1.156774e-09 1.110292e-09
700 1.490994e-09 1.176214e-09
800 1.399189e-09 1.163705e-09
900 1.125521e-09 1.070834e-09
1000 1.517947e-09 1.099748e-09
1100 1.347685e-09 1.464948e-09
1200 1.437535e-09 1.129206e-09
1300 2.319475e-09 1.328544e-09
1400 5.188417e-09 3.125614e-09
1500 2.605655e-09 1.736140e-09
1600 5.250663e-09 1.302334e-09
1700 5.790478e-09 1.337977e-09
1800 8.286016e-09 1.321148e-09
2000 6.833117e-09 1.291707e-09
Plots#
piv_diff = df.pivot(index="dim", columns="direction", values="diff")
piv_time = df.pivot(index="dim", columns="direction", values="time")
fig, ax = plt.subplots(1, 3, figsize=(12, 6))
piv.plot(ax=ax[0], logx=True, title="Comparison between two summation")
piv_diff.plot(ax=ax[1], logx=True, logy=True, title="Summation errors")
piv_time.plot(ax=ax[2], logx=True, logy=True, title="Total time")
fig.savefig("plot_bench_cpu_vector_sum.png")
~/.local/lib/python3.10/site-packages/pandas/plotting/_matplotlib/core.py:741: UserWarning: Data has no positive values, and therefore cannot be log-scaled.
labels = axis.get_majorticklabels() + axis.get_minorticklabels()
The summation by rows is much faster as expected. That explains why it is usually more efficient to transpose the first matrix before a matrix multiplication.
Total running time of the script: ( 0 minutes 18.393 seconds)