Note
Go to the end to download the full example code.
Measuring performance of TfIdfVectorizer¶
The banchmark measures the performance of a TfIdfVectizer along two parameters, the vocabulary size, the batch size whether. It measures the benefit of using sparse implementation through the computation time and the memory peak.
A simple model¶
We start with a model including only one node TfIdfVectorizer. It only contains unigram. The model processes only sequences of 10 integers. The sparsity of the results is then 10 divided by the size of vocabulary.
import gc
import time
import itertools
from typing import Tuple
import numpy as np
import pandas
from onnx import ModelProto
from onnx.helper import make_attribute
from tqdm import tqdm
from onnxruntime import InferenceSession, SessionOptions
from onnx_extended.ext_test_case import measure_time, unit_test_going
from onnx_extended.memory_peak import start_spying_on
from onnx_extended.reference import CReferenceEvaluator
from onnx_extended.ortops.optim.cpu import get_ort_ext_libs
from onnx_extended.plotting.benchmark import vhistograms
def make_onnx(n_words: int) -> ModelProto:
from skl2onnx.common.data_types import Int64TensorType, FloatTensorType
from skl2onnx.algebra.onnx_ops import OnnxTfIdfVectorizer
# from onnx_array_api.light_api import start
# onx = (
# start(opset=19, opsets={"ai.onnx.ml": 3})
# .vin("X", elem_type=TensorProto.INT64)
# .ai.onnx.TfIdfVectorizer(
# ...
# )
# .rename(Y)
# .vout(elem_type=TensorProto.FLOAT)
# .to_onnx()
# )
onx = OnnxTfIdfVectorizer(
"X",
mode="TF",
min_gram_length=1,
max_gram_length=1,
max_skip_count=0,
ngram_counts=[0],
ngram_indexes=np.arange(n_words).tolist(),
pool_int64s=np.arange(n_words).tolist(),
output_names=["Y"],
).to_onnx(inputs=[("X", Int64TensorType())], outputs=[("Y", FloatTensorType())])
# .rename(Y)
# .vout(elem_type=TensorProto.FLOAT)
# .to_onnx()
# )
return onx
onx = make_onnx(7)
ref = CReferenceEvaluator(onx)
got = ref.run(None, {"X": np.array([[0, 1], [2, 3]], dtype=np.int64)})
print(got)
[array([[1., 1., 0., 0., 0., 0., 0.],
[0., 0., 1., 1., 0., 0., 0.]], dtype=float32)]
It works as expected. Let’s now compare the execution with onnxruntime for different batch size and vocabulary size.
Benchmark¶
def make_sessions(
onx: ModelProto,
) -> Tuple[InferenceSession, InferenceSession, InferenceSession]:
# first: onnxruntime
ref = InferenceSession(onx.SerializeToString(), providers=["CPUExecutionProvider"])
# second: custom kernel equivalent to the onnxruntime implementation
for node in onx.graph.node:
if node.op_type == "TfIdfVectorizer":
node.domain = "onnx_extended.ortops.optim.cpu"
# new_add = make_attribute("sparse", 1)
# node.attribute.append(new_add)
d = onx.opset_import.add()
d.domain = "onnx_extended.ortops.optim.cpu"
d.version = 1
r = get_ort_ext_libs()
opts = SessionOptions()
opts.register_custom_ops_library(r[0])
cus = InferenceSession(
onx.SerializeToString(), opts, providers=["CPUExecutionProvider"]
)
# third: with sparse
for node in onx.graph.node:
if node.op_type == "TfIdfVectorizer":
new_add = make_attribute("sparse", 1)
node.attribute.append(new_add)
cussp = InferenceSession(
onx.SerializeToString(), opts, providers=["CPUExecutionProvider"]
)
return ref, cus, cussp
if unit_test_going():
vocabulary_sizes = [10, 20]
batch_sizes = [5, 10]
else:
vocabulary_sizes = [100, 1000, 5000, 10000]
batch_sizes = [1, 10, 500, 1000, 2000]
confs = list(itertools.product(vocabulary_sizes, batch_sizes))
data = []
for voc_size, batch_size in tqdm(confs):
onx = make_onnx(voc_size)
ref, cus, sparse = make_sessions(onx)
gc.collect()
feeds = dict(
X=(np.arange(batch_size * 10) % voc_size)
.reshape((batch_size, -1))
.astype(np.int64)
)
# sparse
p = start_spying_on(delay=0.0001)
sparse.run(None, feeds)
obs = measure_time(
lambda sparse=sparse, feeds=feeds: sparse.run(None, feeds), max_time=1
)
mem = p.stop()
obs["peak"] = mem["cpu"].max_peak - mem["cpu"].begin
obs["name"] = "sparse"
obs.update(dict(voc_size=voc_size, batch_size=batch_size))
data.append(obs)
time.sleep(0.1)
# reference
p = start_spying_on(delay=0.0001)
ref.run(None, feeds)
obs = measure_time(lambda ref=ref, feeds=feeds: ref.run(None, feeds), max_time=1)
mem = p.stop()
obs["peak"] = mem["cpu"].max_peak - mem["cpu"].begin
obs["name"] = "ref"
obs.update(dict(voc_size=voc_size, batch_size=batch_size))
data.append(obs)
time.sleep(0.1)
# custom
p = start_spying_on(delay=0.0001)
cus.run(None, feeds)
obs = measure_time(lambda cus=cus, feeds=feeds: cus.run(None, feeds), max_time=1)
mem = p.stop()
obs["peak"] = mem["cpu"].max_peak - mem["cpu"].begin
obs["name"] = "custom"
obs.update(dict(voc_size=voc_size, batch_size=batch_size))
data.append(obs)
time.sleep(0.1)
del sparse
del cus
del ref
del feeds
df = pandas.DataFrame(data)
df["time"] = df["average"]
df.to_csv("plot_op_tfidfvectorizer_sparse.csv", index=False)
print(df.head())
0%| | 0/20 [00:00<?, ?it/s]
5%|▌ | 1/20 [00:03<01:15, 3.98s/it]
10%|█ | 2/20 [00:08<01:12, 4.04s/it]
15%|█▌ | 3/20 [00:12<01:08, 4.02s/it]
20%|██ | 4/20 [00:16<01:08, 4.29s/it]
25%|██▌ | 5/20 [00:20<01:01, 4.11s/it]
30%|███ | 6/20 [00:24<00:57, 4.14s/it]
35%|███▌ | 7/20 [00:28<00:52, 4.02s/it]
40%|████ | 8/20 [00:32<00:48, 4.05s/it]
45%|████▌ | 9/20 [00:36<00:44, 4.02s/it]
50%|█████ | 10/20 [00:41<00:41, 4.19s/it]
55%|█████▌ | 11/20 [00:45<00:37, 4.15s/it]
60%|██████ | 12/20 [00:49<00:32, 4.05s/it]
65%|██████▌ | 13/20 [00:53<00:29, 4.15s/it]
70%|███████ | 14/20 [00:57<00:25, 4.20s/it]
75%|███████▌ | 15/20 [01:01<00:20, 4.12s/it]
80%|████████ | 16/20 [01:05<00:16, 4.10s/it]
85%|████████▌ | 17/20 [01:09<00:12, 4.09s/it]
90%|█████████ | 18/20 [01:14<00:08, 4.22s/it]
95%|█████████▌| 19/20 [01:18<00:04, 4.20s/it]
100%|██████████| 20/20 [01:22<00:00, 4.17s/it]
100%|██████████| 20/20 [01:22<00:00, 4.13s/it]
average deviation min_exec ... voc_size batch_size time
0 0.000007 7.581871e-07 0.000007 ... 100 1 0.000007
1 0.000006 8.600416e-08 0.000006 ... 100 1 0.000006
2 0.000006 5.242533e-07 0.000005 ... 100 1 0.000006
3 0.000013 3.670098e-07 0.000012 ... 100 10 0.000013
4 0.000019 5.916206e-06 0.000015 ... 100 10 0.000019
[5 rows x 14 columns]
Processing time¶
piv = pandas.pivot_table(
df, index=["voc_size", "name"], columns="batch_size", values="average"
)
print(piv)
batch_size 1 10 500 1000 2000
voc_size name
100 custom 0.000006 0.000006 0.000023 0.000054 0.000050
ref 0.000006 0.000019 0.000027 0.000051 0.000076
sparse 0.000007 0.000013 0.000317 0.000523 0.001015
1000 custom 0.000005 0.000007 0.000054 0.000085 0.000181
ref 0.000006 0.000014 0.000077 0.000150 0.000291
sparse 0.000006 0.000012 0.000299 0.000599 0.001170
5000 custom 0.000006 0.000010 0.000200 0.000389 0.000837
ref 0.000008 0.000017 0.000286 0.000566 0.001150
sparse 0.000006 0.000011 0.000295 0.000557 0.001005
10000 custom 0.000006 0.000014 0.000361 0.000788 0.002377
ref 0.000010 0.000022 0.000572 0.001207 0.002468
sparse 0.000006 0.000011 0.000263 0.000638 0.001057
Memory peak¶
It is always difficult to estimate. A second process is started to measure the physical memory peak during the execution every ms. The figures is the difference between this peak and the memory when the measurement began.
piv = pandas.pivot_table(
df, index=["voc_size", "name"], columns="batch_size", values="peak"
)
print(piv / 2**20)
batch_size 1 10 500 1000 2000
voc_size name
100 custom 0.000000 0.000000 0.000000 0.000000 0.003906
ref 0.000000 0.000000 0.011719 0.000000 0.000000
sparse 0.019531 0.000000 0.035156 0.000000 0.000000
1000 custom 0.000000 0.000000 0.152344 0.000000 0.000000
ref 0.000000 0.007812 0.007812 0.003906 0.011719
sparse 0.000000 0.000000 0.000000 0.000000 0.000000
5000 custom 0.000000 0.000000 9.042969 20.640625 39.378906
ref 0.000000 0.000000 0.015625 0.015625 38.640625
sparse 0.000000 0.000000 0.000000 0.000000 0.000000
10000 custom 0.000000 0.000000 20.640625 39.378906 77.382812
ref 0.000000 0.000000 0.015625 38.640625 77.378906
sparse 0.000000 0.000000 0.000000 0.000000 0.000000
Graphs¶
ax = vhistograms(df)
fig = ax[0, 0].get_figure()
fig.savefig("plot_op_tfidfvectorizer_sparse.png")
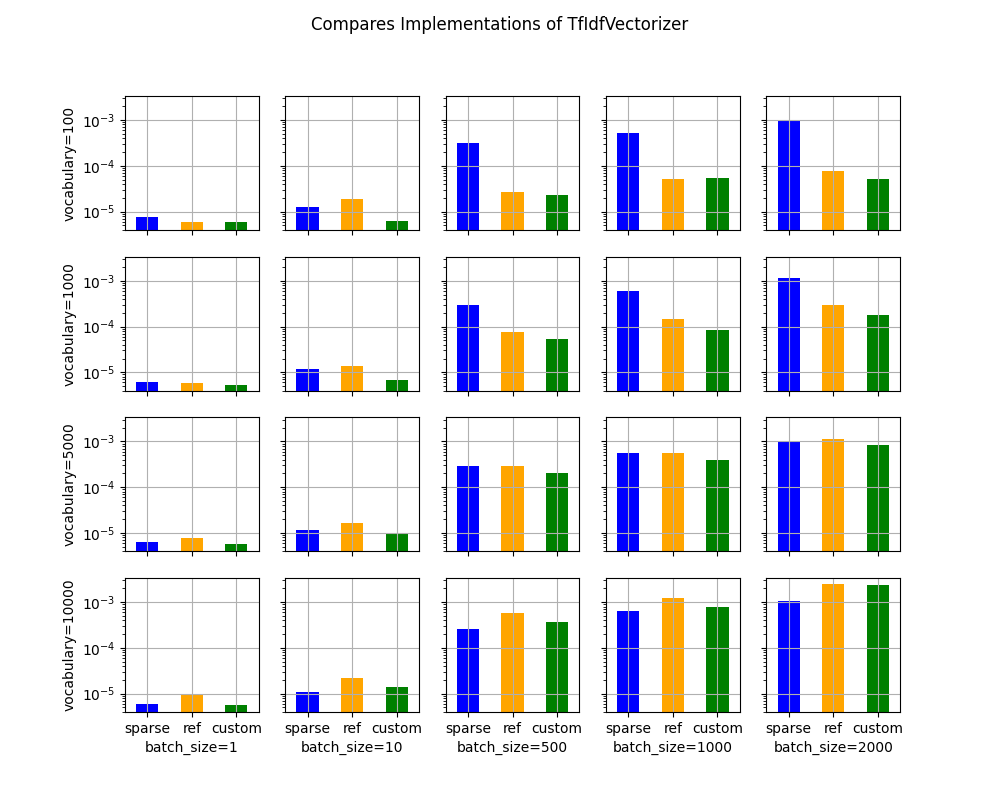
Take away¶
Sparse works better when the sparsity is big enough and the batch size as well.
Total running time of the script: (1 minutes 27.466 seconds)