Note
Go to the end to download the full example code.
LogisticRegression and Clustering¶
A logistic regression implements a convex partition of the features spaces. A clustering algorithm applied before the trainer modifies the feature space in way the partition is not necessarily convex in the initial features. Let’s see how.
A dummy datasets and not convex¶
import numpy
import pandas
import matplotlib.pyplot as plt
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from mlinsights.mlmodel import ClassifierAfterKMeans
Xs = []
Ys = []
n = 20
for i in range(5):
for j in range(4):
x1 = numpy.random.rand(n) + i * 1.1
x2 = numpy.random.rand(n) + j * 1.1
Xs.append(numpy.vstack([x1, x2]).T)
cl = numpy.random.randint(0, 4)
Ys.extend([cl for i in range(n)])
X = numpy.vstack(Xs)
Y = numpy.array(Ys)
X.shape, Y.shape, set(Y)
((400, 2), (400,), {np.int64(0), np.int64(1), np.int64(2), np.int64(3)})
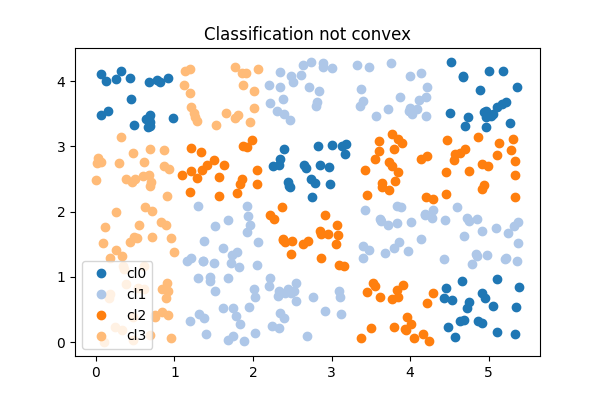
Text(0.5, 1.0, 'Classification not convex')
One function to plot classification in 2D¶
def draw_border(
clr,
X,
y,
fct=None,
incx=1,
incy=1,
figsize=None,
border=True,
clusters=None,
ax=None,
):
# see https://sashat.me/2017/01/11/list-of-20-simple-distinct-colors/
# https://matplotlib.org/examples/color/colormaps_reference.html
h = 0.02 # step size in the mesh
# Plot the decision boundary. For that, we will assign a color to each
# point in the mesh [x_min, x_max]x[y_min, y_max].
x_min, x_max = X[:, 0].min() - incx, X[:, 0].max() + incx
y_min, y_max = X[:, 1].min() - incy, X[:, 1].max() + incy
xx, yy = numpy.meshgrid(
numpy.arange(x_min, x_max, h), numpy.arange(y_min, y_max, h)
)
if fct is None:
Z = clr.predict(numpy.c_[xx.ravel(), yy.ravel()])
else:
Z = fct(clr, numpy.c_[xx.ravel(), yy.ravel()])
# Put the result into a color plot
cmap = plt.cm.tab20
Z = Z.reshape(xx.shape)
if ax is None:
_fig, ax = plt.subplots(1, 1, figsize=figsize or (4, 3))
ax.pcolormesh(xx, yy, Z, cmap=cmap)
# Plot also the training points
ax.scatter(X[:, 0], X[:, 1], c=y, edgecolors="k", cmap=cmap)
ax.set_xlabel("Sepal length")
ax.set_ylabel("Sepal width")
ax.set_xlim(xx.min(), xx.max())
ax.set_ylim(yy.min(), yy.max())
# Plot clusters
if clusters is not None:
mat = []
ym = []
for k, v in clusters.items():
mat.append(v.cluster_centers_)
ym.extend(k for i in range(v.cluster_centers_.shape[0]))
cx = numpy.vstack(mat)
ym = numpy.array(ym)
ax.scatter(cx[:, 0], cx[:, 1], c=ym, edgecolors="y", cmap=cmap, s=300)
return ax
Logistic Regression¶
clr = LogisticRegression(solver="lbfgs")
clr.fit(X, Y)
ax = draw_border(clr, X, Y, incx=1, incy=1, figsize=(6, 4), border=False)
ax.set_title("Logistic Regression")
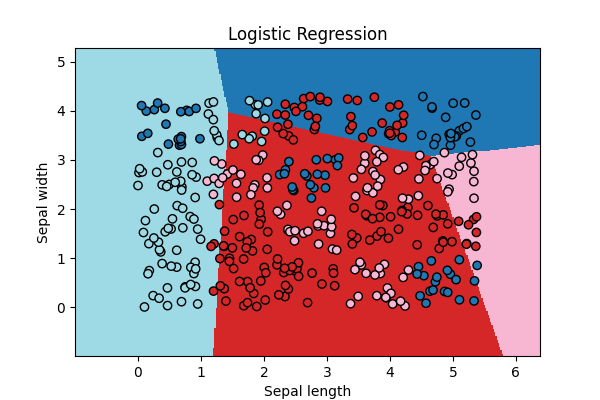
Text(0.5, 1.0, 'Logistic Regression')
Not quite close!
Logistic Regression and k-means¶
clk = ClassifierAfterKMeans(e_solver="lbfgs")
clk.fit(X, Y)
~/vv/this312/lib/python3.12/site-packages/sklearn/linear_model/_logistic.py:473: ConvergenceWarning: lbfgs failed to converge after 100 iteration(s) (status=1):
STOP: TOTAL NO. OF ITERATIONS REACHED LIMIT
Increase the number of iterations to improve the convergence (max_iter=100).
You might also want to scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
Please also refer to the documentation for alternative solver options:
https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression
n_iter_i = _check_optimize_result(
The centers of the first k-means:
clk.clus_[0].cluster_centers_
array([[2.73229243, 3.36426616],
[4.84500873, 0.52154296]])
ax = draw_border(
clk, X, Y, incx=1, incy=1, figsize=(6, 4), border=False, clusters=clk.clus_
)
ax.set_title("Logistic Regression and K-Means - 2 clusters per class")
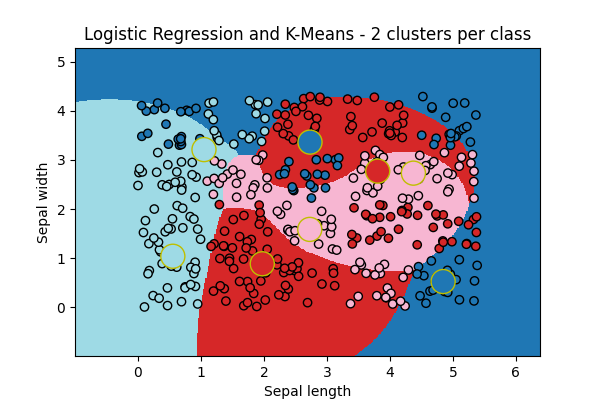
Text(0.5, 1.0, 'Logistic Regression and K-Means - 2 clusters per class')
The big cricles are the centers of the k-means fitted for each class. It look better!
Variation¶
ax = draw_border(
clk, X, Y, incx=1, incy=1, figsize=(6, 4), border=False, clusters=clk.clus_
)
ax.set_title("Logistic Regression and K-Means - 8 clusters per class")
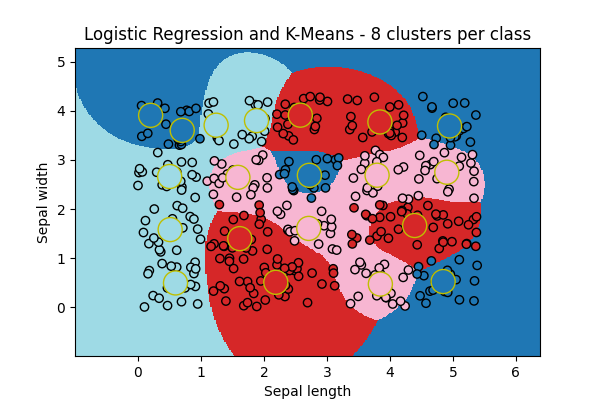
Text(0.5, 1.0, 'Logistic Regression and K-Means - 8 clusters per class')
Random Forest¶
# The random forest works without any clustering as expected.
rf = RandomForestClassifier(n_estimators=20)
rf.fit(X, Y)
ax = draw_border(rf, X, Y, incx=1, incy=1, figsize=(6, 4), border=False)
ax.set_title("Random Forest")
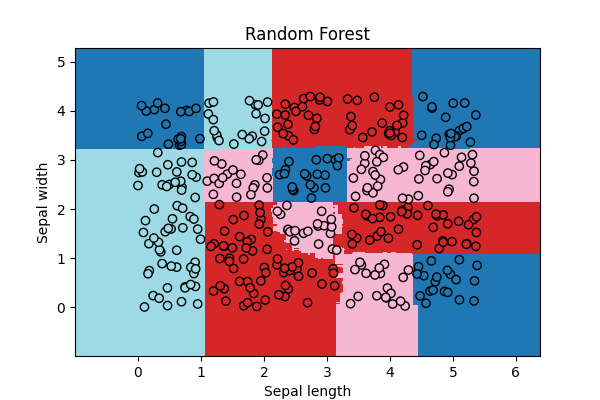
Text(0.5, 1.0, 'Random Forest')
Total running time of the script: (0 minutes 1.412 seconds)